| Line 124: | Line 124: | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/HP/Silver">Silver</a> </li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/HP/Silver">Silver</a> </li> | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/HP/Gold">Gold</a> </li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/HP/Gold">Gold</a> </li> | ||
| + | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Medal_Criteria">Medal Criteria</a> </li> | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/Integrated_Practices">Integrated Practices</a> </li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Integrated_Practices">Integrated Practices</a> </li> | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/Engagement">Engagement</a> </li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Engagement">Engagement</a> </li> | ||
| Line 138: | Line 139: | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/Demonstrate">Demonstrate</a></li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Demonstrate">Demonstrate</a></li> | ||
<li><a href="https://2016.igem.org/Team:Edinburgh_UG/Notebook">Notebook</a></li> | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Notebook">Notebook</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Protocols">Protocols</a></li> | ||
| + | <li><a href="https://2016.igem.org/Team:Edinburgh_UG/Limitations">Limitations</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
| Line 186: | Line 189: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
| Line 277: | Line 279: | ||
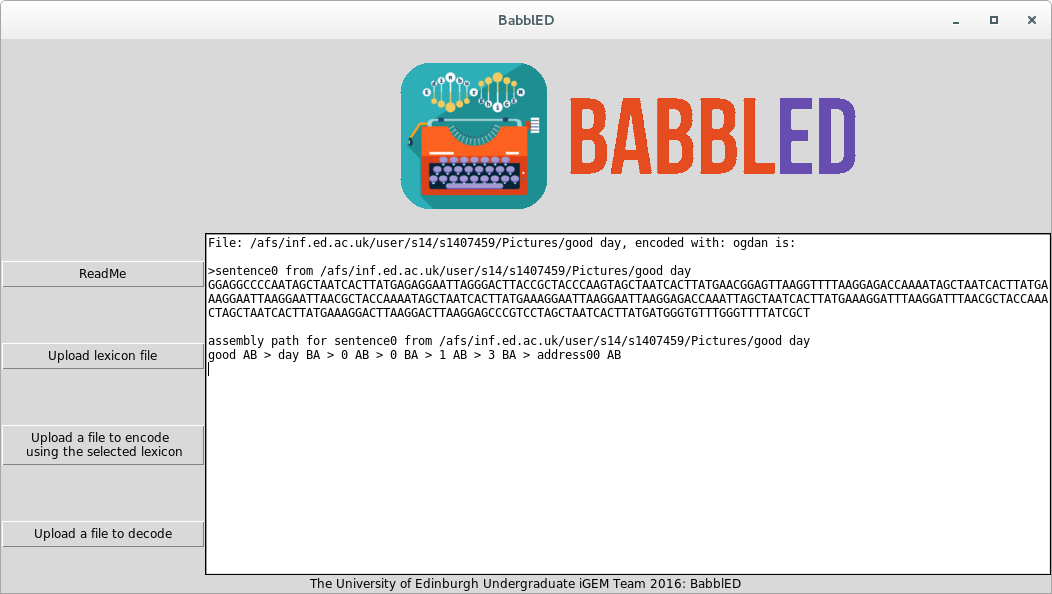
<centre style="font-size:160%;">Python is high-level, general purpose language designed for quick creation of highly readable code. As a result of libraries such as BioPython it has becoming increasingly used in Bioinformatics and Computational Biology in recent years this made it the obvious choice for the BabblED software. We made the decision to use Python 2.7 to increase accessibility as this is the Python version commonly pre-installed on Linux distributions such as Ubuntu.</centre> | <centre style="font-size:160%;">Python is high-level, general purpose language designed for quick creation of highly readable code. As a result of libraries such as BioPython it has becoming increasingly used in Bioinformatics and Computational Biology in recent years this made it the obvious choice for the BabblED software. We made the decision to use Python 2.7 to increase accessibility as this is the Python version commonly pre-installed on Linux distributions such as Ubuntu.</centre> | ||
<p> </p> | <p> </p> | ||
| − | <centre style="font-size:160%;">Why avoid a web app?</centre> | + | <centre style="font-size:160%;">Why avoid a web app?</centre> |
<p> </p> | <p> </p> | ||
<centre style="font-size:160%;">When developing our software we carefully considered how it should be deployed. Should we create a web app using Django (the commonly used Python web framework)? Or simply make all our code easily downloadable with documentation on how to run it? Despite the minor drawbacks in accessibility we eventually settled on the later option for two main reasons. Firstly BabblED was designed for encoding large amounts of archival data - such large amounts that sending these volumes over the web would be very impracticable. Secondly having put a lot of work into the data security angle of the project we deemed that the web app approach had a much higher chance to introduce unforeseen security flaws that we knew (not being computer security experts) we would not have the capabilities to protect against.</centre> | <centre style="font-size:160%;">When developing our software we carefully considered how it should be deployed. Should we create a web app using Django (the commonly used Python web framework)? Or simply make all our code easily downloadable with documentation on how to run it? Despite the minor drawbacks in accessibility we eventually settled on the later option for two main reasons. Firstly BabblED was designed for encoding large amounts of archival data - such large amounts that sending these volumes over the web would be very impracticable. Secondly having put a lot of work into the data security angle of the project we deemed that the web app approach had a much higher chance to introduce unforeseen security flaws that we knew (not being computer security experts) we would not have the capabilities to protect against.</centre> | ||
Revision as of 22:24, 19 October 2016