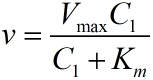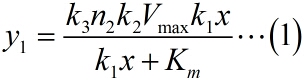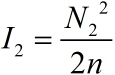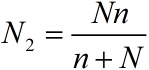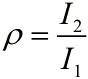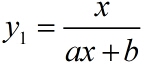Qiuxinyuan12 (Talk | contribs) |
Zhuchushu13 (Talk | contribs) |
||
| Line 393: | Line 393: | ||
| − | <br/><br/><br/>NUDT_CHINA: This page is under | + | <!--标题--> |
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Abstract</span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This | ||
| + | |||
| + | model was created to evaluate the effectiveness of initial design, and offer | ||
| + | |||
| + | guidelines about how the system can (or must) be improved. (You can go PROJECT | ||
| + | |||
| + | page to see more.) The core idea was to simulate the process of producing the | ||
| + | |||
| + | signal which can be detected, and drew a conclusion by obtaining the | ||
| + | |||
| + | relationship between the signal intensity and the concentration of miRNA. | ||
| + | |||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--标题--> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Introduction </span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Mathematical models of two aspects of our project were created, a | ||
| + | |||
| + | RCA model and a signal detection model. | ||
| + | |||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA | ||
| + | |||
| + | model was based on the Michaelis-Menten equation. The relationship between the | ||
| + | |||
| + | concentration of miRNA and the number of stem-loop structures was obtained | ||
| + | |||
| + | through theoretical calculation, and our experimental results was used to | ||
| + | |||
| + | calculate the parameters we introduced previously.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | signal detection model was composed of a probability model and a kinetic | ||
| + | |||
| + | equation of enzymatic reaction, thus the relationship between the number of | ||
| + | |||
| + | stem-loop structures and the signal intensity under the premise of adding a | ||
| + | |||
| + | certain amount of fusion proteins of dCas9 and split-HRP fragments was obtained. | ||
| + | |||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By | ||
| + | |||
| + | integrating the two models, the impacts of the molecule number of proteins on | ||
| + | |||
| + | the signal to noise ratio was theoretically predicted and the trend of the | ||
| + | |||
| + | signal intensity with the change of the concentration of miRNA in our wet-lab | ||
| + | |||
| + | work was explained.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015"> Assumption and Justification | ||
| + | |||
| + | </span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> About model </span></span><hr /> | ||
| + | </h3> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | ||
| + | |||
| + | We assume that miRNA is not degraded throughout the reaction process.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | ||
| + | |||
| + | We assume that the two fusion proteins of dCas9 and split-HRP fragments have the | ||
| + | |||
| + | same ability to combine with the stem-loop structure, and only when two | ||
| + | |||
| + | different proteins next to each other, can they have the ability to catalyze | ||
| + | |||
| + | substrate and produce signal.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | ||
| + | |||
| + | We assume that the number of stem-loop structures in each RCA product is equal | ||
| + | |||
| + | under a certain reaction time. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | ||
| + | We assume that the enzymatic activity remains unchanged with time under the | ||
| + | |||
| + | premise of excessive amount of enzymes or a short-time reaction.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> About the data </span></span><hr | ||
| + | |||
| + | /> | ||
| + | </h3> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> We assume that the data we obtain from wet-lab experiment are | ||
| + | |||
| + | reliable.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> We assume that all the results are trustworthy in the process of | ||
| + | |||
| + | statistical processing and data calculation.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Model </span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015">Notations </span></span><hr /> | ||
| + | </h3> | ||
| + | |||
| + | |||
| + | |||
| + | <!--表格--> | ||
| + | <p> | ||
| + | <table class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" style="border:none;"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>Symbol </span> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>Definition </span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>X</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK7"></a><a name="OLE_LINK6"></a><span>The concentration of</span><span> miRNA (pM)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>C<sub>1</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The concentration of initiated probe (Abbreviated | ||
| + | to iprobe)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>k<sub>1</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK8"></a><span>A constant representing | ||
| + | the scale factor</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>K<sub>m </sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>One of the characteristic constants of | ||
| + | phi29 DNA polymerase</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>V<sub>max</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>One of the characteristic constants of phi29 | ||
| + | DNA polymerase</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>k<sub>2</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>A constant representing the scale factor</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>V</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The initial speed of RCA </span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></sub></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The moles of RCA product</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>n<sub>2</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK10"></a><span>The number of | ||
| + | stem-loop structures in each RCA product</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>n</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The total amount of <a name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>N</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK41"></a><a name="OLE_LINK16"></a><span>The molecule number of fusion | ||
| + | protein of dCas9 and split-HRP fragments</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>k<sub>3</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>A constant representing the scale factor</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>y<sub>1</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The fluorescence intensity of | ||
| + | DNA-dye-complex (RFU)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>N<sub>1</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The molecule number of fusion protein of | ||
| + | dCas9 and split-HRP fragments in the solution</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>N<sub>2</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK45"></a><span>The molecule number | ||
| + | of fusion protein of dCas9 and split-HRP fragments binding with stem-loop | ||
| + | structure</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>k<sub>4</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>A constant representing the scale factor</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span>k<sub>5</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>A constant representing the scale factor</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">ρ</span><span></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>Signal to noise ratio(Abbreviated to | ||
| + | SNR)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">I</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The molecule number of formed intact HRP | ||
| + | proteins </span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">I<sub>1</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The molecule number of formed intact HRP | ||
| + | proteins in the solution</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">I<sub>2</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <a name="OLE_LINK43"></a><span>The molecule number | ||
| + | of formed intact HRP proteins through binding with stem-loop structure</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">t</span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>Reaction time</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <i><span style="font-family:宋体;">y<sub>2</sub></span></i> | ||
| + | </p> | ||
| + | </td> | ||
| + | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | ||
| + | <p class="MsoNormal"> | ||
| + | <span>The signal intensity (OD<sub>450</sub>)</span> | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> The RCA model </span></span><hr | ||
| + | |||
| + | /> | ||
| + | </h3> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, we assume that there is a linear relationship between the | ||
| + | |||
| + | concentration of the initiated probes and miRNA as the combination reaction | ||
| + | |||
| + | between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written | ||
| + | |||
| + | as: | ||
| + | |||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model1.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | ||
| + | |||
| + | RCA is an enzymatic reaction, the reaction equation is written as: iprobe+dNTP→ | ||
| + | |||
| + | iprobe-dN+PPi, under the premise of excessive amount of enzymes and dNTPs, the | ||
| + | |||
| + | relationship between the concentration of iprobe and the initial speed of RCA | ||
| + | |||
| + | can be described by the Michaelis-Menten equation. This can be written as: | ||
| + | |||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model2.jpg|105px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">It is | ||
| + | |||
| + | obvious that there is a linear relationship between the molecule numbers of | ||
| + | |||
| + | iprobe-dN (n<sub>1 </sub>, reflects the molecule numbers of RCA product) and the initial | ||
| + | |||
| + | speed of RCA reaction. Notably, under the premise of excessive amount of | ||
| + | |||
| + | enzymes, the extending of each RCA production is related to the reaction time, | ||
| + | |||
| + | not the concentration of the iprobe. It is a linear relationship when we assume | ||
| + | |||
| + | that the enzymatic activity remains unchanged with time, thus, the number of | ||
| + | |||
| + | stem-loop structures (n<sub>2 </sub>) in each RCA product is stable under the premise of a | ||
| + | |||
| + | certain reaction time. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig1.jpg|500px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--图注--> | ||
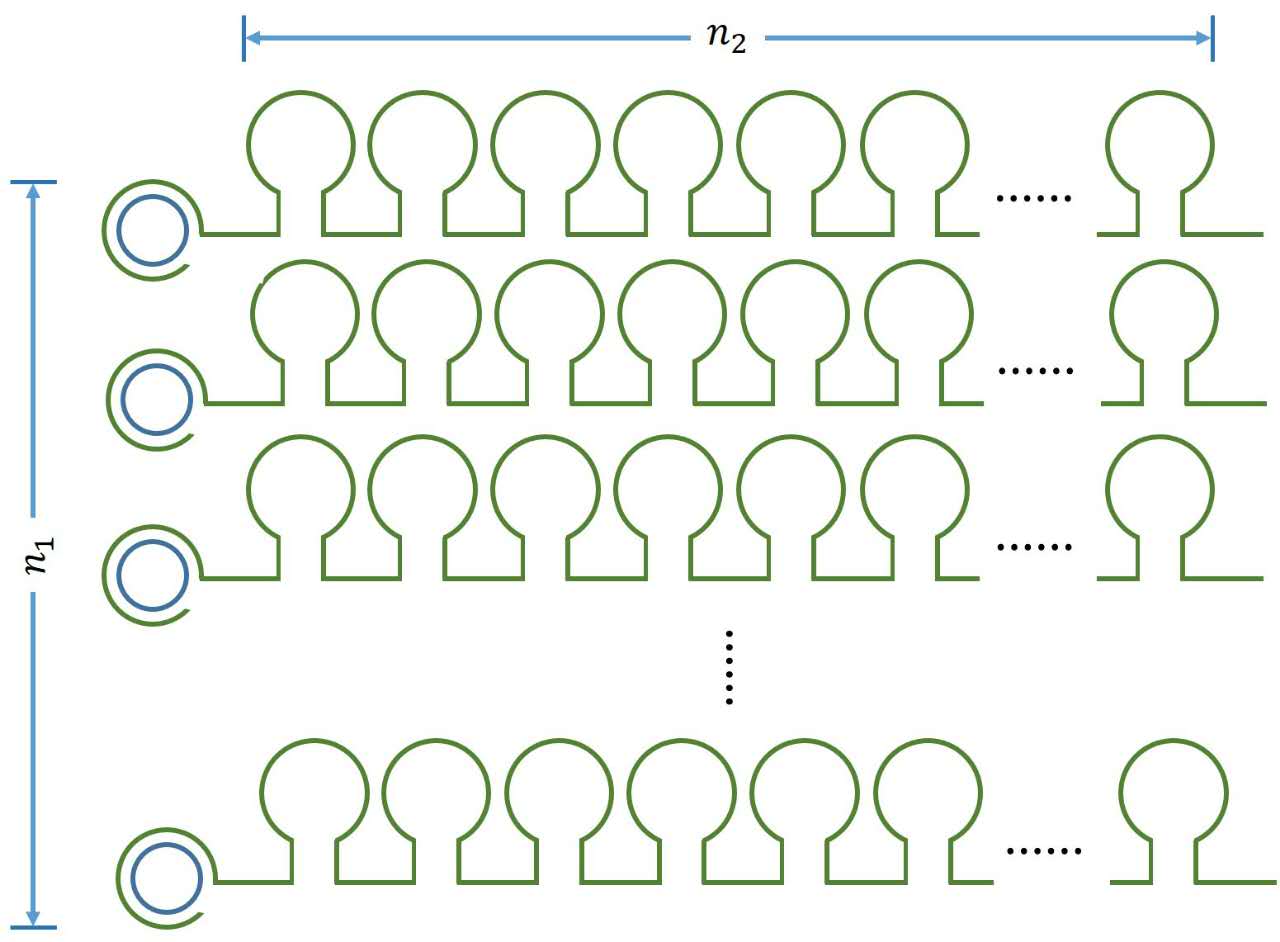
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure 1. Schematic diagram</span></b> | ||
| + | </p> | ||
| + | |||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This | ||
| + | |||
| + | can be written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model3.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model4.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And | ||
| + | |||
| + | then, SYBR Green binds to DNA. The resulting DNA-dye-complex absorbs blue light | ||
| + | |||
| + | (λmax = 497nm) and emits green light (λmax = 520nm). Thus, there is a linear | ||
| + | |||
| + | relationship between the total amount of stem-loop structures and the | ||
| + | |||
| + | fluorescence intensity of DNA-dye-complex. This can be written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model5.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In | ||
| + | |||
| + | summary, the relationship between the concentration of miRNA and the | ||
| + | |||
| + | fluorescence intensity of DNA-dye-complex can be written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model6.jpg|280px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> The signal detection model | ||
| + | |||
| + | </span></span><hr /> | ||
| + | </h3> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Firstly, it is obvious that there is a linear relationship between | ||
| + | |||
| + | the molecule number of formed intact HRP proteins in the solution (consider as | ||
| + | |||
| + | NOISE) and the molecule number of fusion protein s in the solution. This can be | ||
| + | |||
| + | written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model7.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
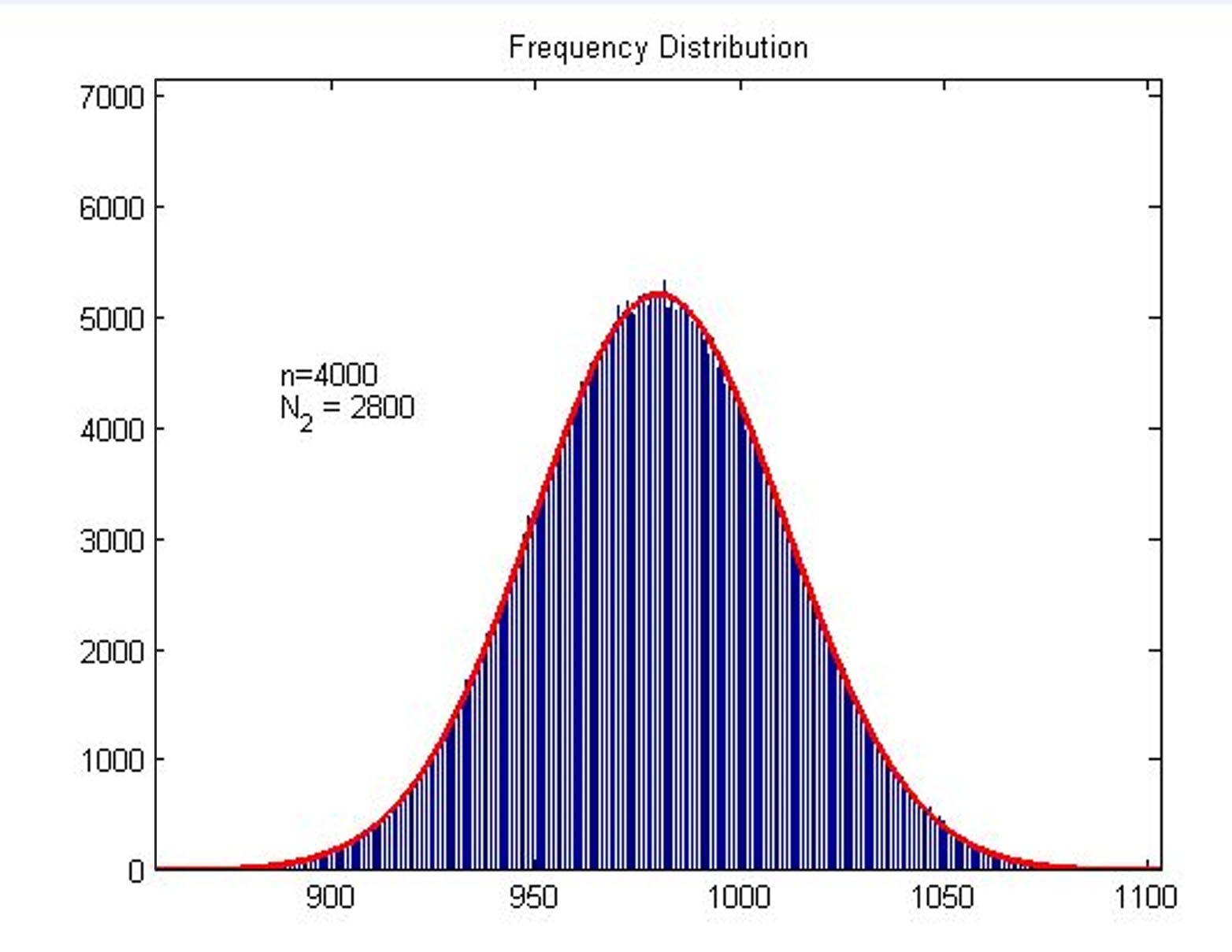
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, | ||
| + | |||
| + | we built a probability model to find the expression of the molecule number of | ||
| + | |||
| + | formed intact HRP proteins (fused with dCas9) through binding with stem-loop | ||
| + | |||
| + | structure (consider as SIGNAL). It is related to the total amount of stem-loop | ||
| + | |||
| + | structures and the molecule number of DNA-bound fusion proteins. We obtain the | ||
| + | |||
| + | result by Monte Carlo method. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig2.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure2. Frequency Distribution</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Plot of | ||
| + | |||
| + | frequency distribution of the molecule number of formed intact HRP proteins | ||
| + | |||
| + | through binding with stem-loop structure. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig3.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
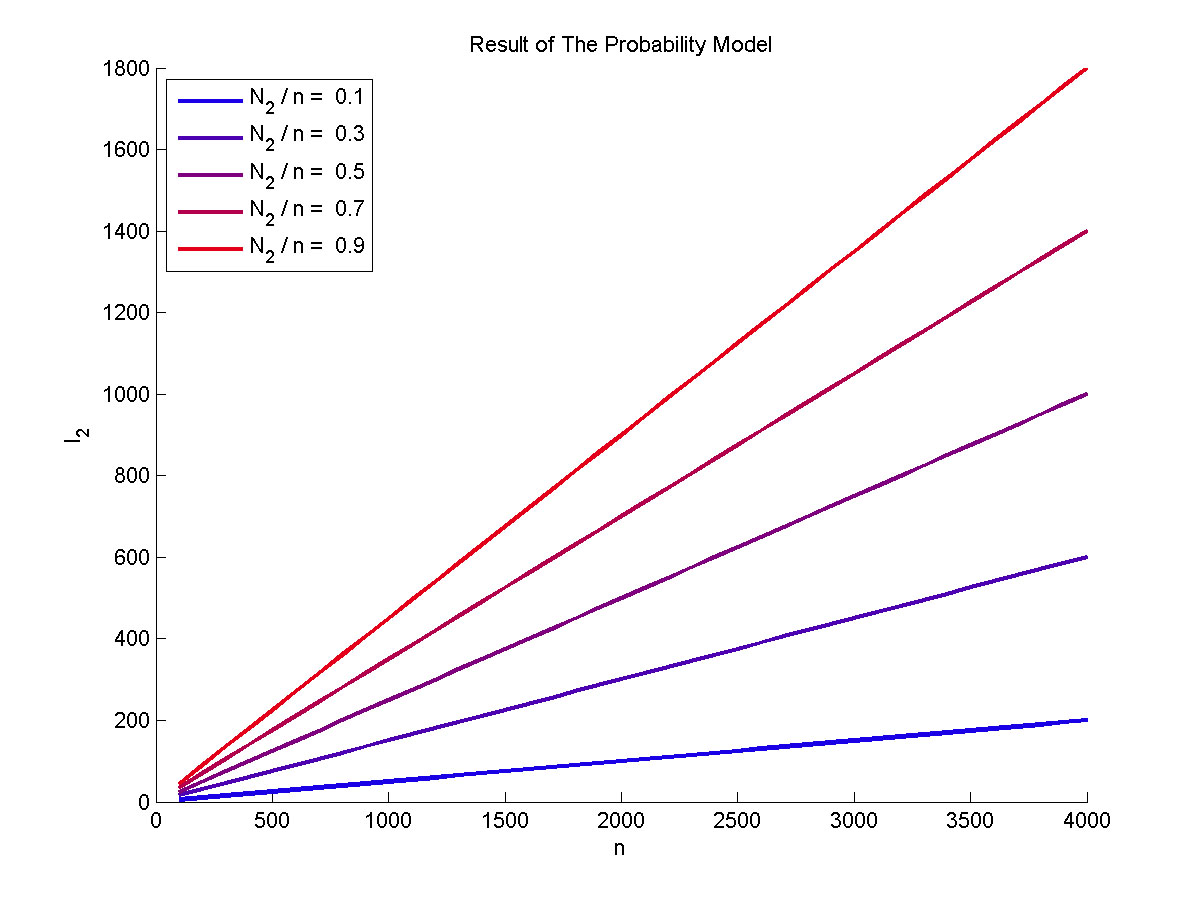
| + | size:18px;">Figure 3. The result of the probability model</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Plot of | ||
| + | |||
| + | the expect value of the molecule number of formed intact proteins against the | ||
| + | |||
| + | total amount of stem-loop structures, carried out at 0.1, 0.3, 0.5, 0.7, 0.9 of | ||
| + | |||
| + | N2 to n ratio. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | result can be written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model8.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for | ||
| + | |||
| + | N2, which refers to the molecule number of fusion proteins binding with stem- | ||
| + | |||
| + | loop structure. The equation is formulated based on the limiting case. The | ||
| + | |||
| + | equation can be written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model9.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And | ||
| + | |||
| + | then, we can obtain the expressions for ρ and I separately.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model10.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model11.jpg|70px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Notably, under the premise of excessive substrates, it is a linear | ||
| + | |||
| + | relationship between the reaction rate and the concentration of enzymes when we | ||
| + | |||
| + | assume that the enzymatic activity remain unchanged with time, thus, we can | ||
| + | |||
| + | obtain the expression of the signal intensity at tth time-step. This is written | ||
| + | |||
| + | as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model12.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In | ||
| + | |||
| + | summary, the relationship between the number of stem-loop structures and the | ||
| + | |||
| + | signal to noise ratio under the premise of adding a certain amount of fusion | ||
| + | |||
| + | proteins is written as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model13.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | relationship between the number of Stem-loop structures and the signal intensity | ||
| + | |||
| + | under the premise of adding a certain amount of fusion proteins is written | ||
| + | |||
| + | as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model14.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> The calculation of the constants | ||
| + | |||
| + | </span></span><hr /> | ||
| + | </h3> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To | ||
| + | |||
| + | simplify the equation (1), the constants could be integrated. As can be seen | ||
| + | |||
| + | from the above table, k1, k2, k3 are constants representing the scale factor, Km | ||
| + | |||
| + | and Vmax are characteristic constants of phi 29 DNA polymerase, and n2, which | ||
| + | |||
| + | refers to the number of stem-loop structures in each RCA product, is stable | ||
| + | |||
| + | under the premise of a certain reaction time. Thus, define two constants, mark | ||
| + | |||
| + | as a and b, and then the equation can be simplified as:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--插入图片--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--model15.jpg|140px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We used | ||
| + | |||
| + | this equation to fit the data points obtained through experiments. (Figure …in | ||
| + | |||
| + | the ….page) The fitting curve is shown below.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig4.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure 4. Fitting curve</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Fitting | ||
| + | |||
| + | curve of fluorescence intensity of DNA-dye-complex (RFU) against the | ||
| + | |||
| + | concentrations of miRNA (Pm). </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">With | ||
| + | |||
| + | regard to the equation (2), we set the parameters based on the experiments as | ||
| + | |||
| + | follows: | ||
| + | k3 =8.35*10-16 (n=y1/k3); k4 =0.038; k5t=5.9*10-12. | ||
| + | </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h3> | ||
| + | <span><span style="color:#7f1015"> Results and Analysis | ||
| + | |||
| + | </span></span><hr /> | ||
| + | </h3> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By | ||
| + | |||
| + | integrating the two models, we can obtain the relationships between the signal | ||
| + | |||
| + | to noise ratio, the signal intensity respectively and the concentration of miRNA | ||
| + | |||
| + | under different additional amount of fusion proteins.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | results are shown below.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig5.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
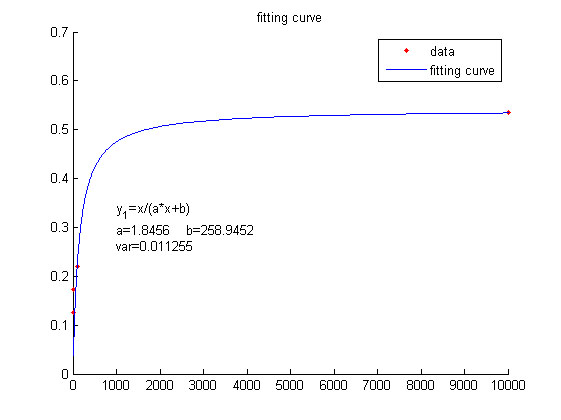
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure 5. The signal intensity (OD450)</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Three- | ||
| + | |||
| + | dimensional map of signal intensity (OD450) against miRNA concentration(pM) and | ||
| + | |||
| + | additional amount of fusion proteins. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">There | ||
| + | |||
| + | exists a monotonous relation between the concentration of miRNA and the signal | ||
| + | |||
| + | intensity when the value of the molecule number of fusion proteins is relatively | ||
| + | |||
| + | large. While the relationship does not hold when the value of the molecule | ||
| + | |||
| + | number of fusion proteins is relatively small. And the signal intensity | ||
| + | |||
| + | increases as the value of the molecule number of fusion proteins | ||
| + | |||
| + | increases.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | trend of the signal intensity with the change of the concentration of miRNA is | ||
| + | |||
| + | consistent with that we obtained in our wet-lab work.(…….. in the result | ||
| + | |||
| + | page)</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig6.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
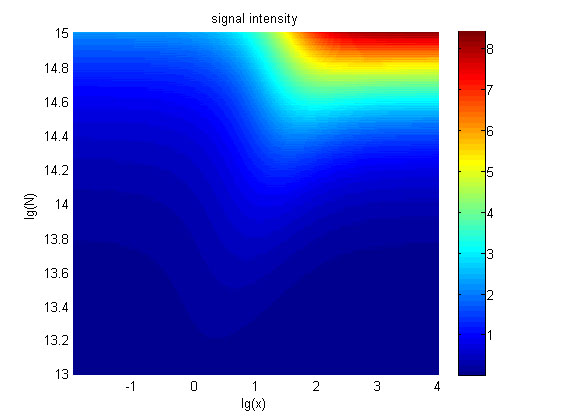
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure 6. The result of signal to noise ratio</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Three- | ||
| + | |||
| + | dimensional map of signal to noise ratio against miRNA concentration (pM) and | ||
| + | |||
| + | additional amount of fusion proteins.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | signal-to-noise ratio decreases as the value of the molecule number of fusion | ||
| + | |||
| + | proteins increases. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
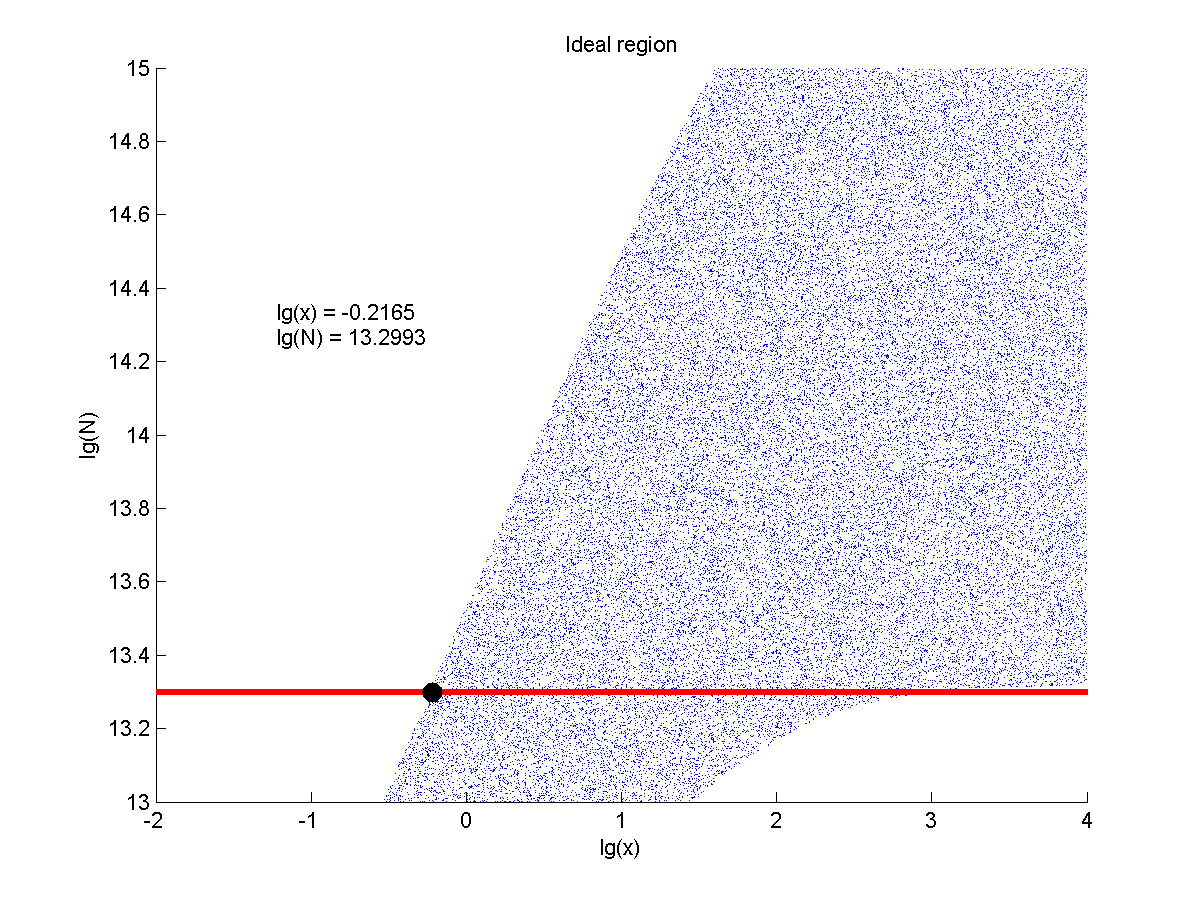
| + | size:18px;">Furthermore, we consider that it is conducive to signal detection | ||
| + | |||
| + | when the values of signal intensity and signal to noise ratio are both greater | ||
| + | |||
| + | than two. We obtain the ideal region through calculation, which is shown as | ||
| + | |||
| + | below. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--图和图注--> | ||
| + | </br> | ||
| + | </html> | ||
| + | [[File:T--NUDT_CHINA--modelfig7.jpg|700px|center]] | ||
| + | <html> | ||
| + | </br> | ||
| + | <p> | ||
| + | <b><span style="line-height:2;font-family:Perpetua;font- | ||
| + | |||
| + | size:18px;">Figure 7. Ideal region</span></b> | ||
| + | </p> | ||
| + | <p> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:16px;">Region | ||
| + | |||
| + | of qualified logarithm of concentrations of miRNA (pM) and logarithm of the | ||
| + | |||
| + | additional amount of fusion proteins.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When | ||
| + | |||
| + | the value of N is set to 1013.3, the range of the value of x contained in the | ||
| + | |||
| + | ideal region is the largest, which means when the value of the molecule number | ||
| + | |||
| + | of fusion proteins is 1013.3, the concentration range of miRNA contained in the | ||
| + | |||
| + | ideal region is the largest. </span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Conclusion </span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Based | ||
| + | |||
| + | on our simulation, we came to the conclusion that:</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | calculated trend of the signal intensity with the change of the concentration of | ||
| + | |||
| + | miRNA from our model is consistent with the trend we obtained in our laboratory | ||
| + | |||
| + | work.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | value of the molecule number of fusion protein of dCas9 and split-HRP fragments | ||
| + | |||
| + | not only affects the signal-to-noise ratio, but also the signal intensity. So we | ||
| + | |||
| + | need to weigh its impact on both to select the optimal solution.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The | ||
| + | |||
| + | concentration range of miRNA contained in the ideal region is the largest when | ||
| + | |||
| + | we set the value of the molecule number of fusion proteins to be 1013.3, we | ||
| + | |||
| + | optimize the value of the addition amount of fusion proteins in our wet-lab work | ||
| + | |||
| + | based on it.</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | <!--标题--> | ||
| + | <h2> | ||
| + | <span><span style="color:#7f1015">Reference </span></span><hr /> | ||
| + | </h2> | ||
| + | |||
| + | <!--正文--> | ||
| + | <p style="text-indent:22pt;"> | ||
| + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1 | ||
| + | |||
| + | Deng, R. et al. Toehold-initiated rolling circle amplification for visualizing | ||
| + | |||
| + | individual microRNAs in situ in single cells. Angew Chem Int Ed Engl 53, 2389- | ||
| + | |||
| + | 2393, doi:10.1002/anie.201309388 (2014).</span> | ||
| + | </p> | ||
| + | </br> | ||
| + | |||
| + | |||
| + | |||
Revision as of 02:46, 15 October 2016