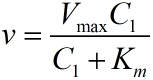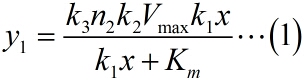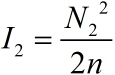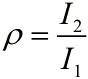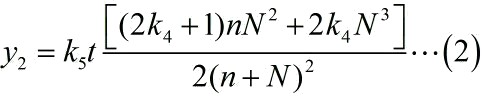Zhuchushu13 (Talk | contribs) |
Zhuchushu13 (Talk | contribs) |
||
| Line 400: | Line 400: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This model is created to evaluate the effectiveness of initial design, and offers guidelines about how the system can (or must) be improved. (You can go PROJECT page to see more.) The core idea is to simulate the process of producing the signal which can be detected, and draw a conclusion by obtaining the relationship between the signal intensity and the concentration of miRNA. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This |
| + | |||
| + | model is created to evaluate the effectiveness of initial design, and offers | ||
| + | |||
| + | guidelines about how the system can (or must) be improved. (You can go PROJECT | ||
| + | |||
| + | page to see more.) The core idea is to simulate the process of producing the | ||
| + | |||
| + | signal which can be detected, and draw a conclusion by obtaining the | ||
| + | |||
| + | relationship between the signal intensity and the concentration of miRNA. | ||
| + | |||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 412: | Line 424: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We create mathematical models of two aspects of our project, a RCA model and a signal detection model.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | create mathematical models of two aspects of our project, a RCA model and a | ||
| + | |||
| + | signal detection model.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 419: | Line 435: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA model is based on the Michaelis-Menten equation. We can obtain the relationship between the concentration of miRNA and the number of stem-loop structures through theoretical calculation, and we use our experimental results to calculate the parameters we introduced previously.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The RCA |
| + | |||
| + | model is based on the Michaelis-Menten equation. We can obtain the relationship | ||
| + | |||
| + | between the concentration of miRNA and the number of stem-loop structures | ||
| + | |||
| + | through theoretical calculation, and we use our experimental results to | ||
| + | |||
| + | calculate the parameters we introduced previously.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 426: | Line 450: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the signal detection model, we combine a probability model with the kinetic equation of enzymatic reaction, so we can obtain the relationship between the number of stem-loop structures and the signal intensity under the premise of adding a certain amount of the fusion proteins of dCas9 and split-HRP fragments. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In the |
| + | |||
| + | signal detection model, we combine a probability model with the kinetic equation | ||
| + | |||
| + | of enzymatic reaction, so we can obtain the relationship between the number of | ||
| + | |||
| + | stem-loop structures and the signal intensity under the premise of adding a | ||
| + | |||
| + | certain amount of the fusion proteins of dCas9 and split-HRP fragments. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 433: | Line 465: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By integrating the two models, we can theoretically predict the impacts of the molecule number of proteins on the signal to noise ratio and explained the trend of the signal intensity with the change of the concentration of miRNA in our wet-lab work. | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By |
| + | |||
| + | integrating the two models, we can theoretically predict the impacts of the | ||
| + | |||
| + | molecule number of proteins on the signal to noise ratio and explained the trend | ||
| + | |||
| + | of the signal intensity with the change of the concentration of miRNA in our | ||
| + | |||
| + | wet-lab work. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 441: | Line 481: | ||
<!--标题--> | <!--标题--> | ||
<h2> | <h2> | ||
| − | <span><span style="color:#7f1015"> Assumption and Justification </span></span><hr /> | + | <span><span style="color:#7f1015"> Assumption and Justification |
| + | |||
| + | </span></span><hr /> | ||
</h2> | </h2> | ||
| Line 452: | Line 494: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that miRNA is not degraded throughout the reaction process.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 459: | Line 503: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that the two fusion proteins of dCas9 and split-HRP fragments have the | ||
| + | |||
| + | same ability to combine with the stem-loop structure, and only when two | ||
| + | |||
| + | different proteins next to each other, can they have the ability to catalyze | ||
| + | |||
| + | substrate and produce signal.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 466: | Line 518: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that the number of stem-loop structures in each RCA product is equal | ||
| + | |||
| + | under a certain reaction time. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 473: | Line 529: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that the enzymatic activity remains unchanged with time under the premise | ||
| + | |||
| + | of excessive amount of enzymes or a short-time reaction.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 481: | Line 541: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> About the data </span></span><hr /> | + | <span><span style="color:#7f1015"> About the data </span></span><hr |
| + | |||
| + | /> | ||
</h3> | </h3> | ||
| Line 487: | Line 549: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that the data we obtain from wet-lab experiment are reliable.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 494: | Line 558: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;"> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We |
| + | |||
| + | assume that all the results are trustworthy in the process of statistical | ||
| + | |||
| + | processing and data calculation.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 512: | Line 580: | ||
<!--表格--> | <!--表格--> | ||
| − | <table class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" style="border:none;"> | + | <table class="MsoTableGrid" border="1" cellspacing="0" cellpadding="0" |
| + | |||
| + | style="border:none;"> | ||
<tbody> | <tbody> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>Symbol </span> | <span>Symbol </span> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>Definition </span> | <span>Definition </span> | ||
| Line 527: | Line 601: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>x</span></i> | <i><span>x</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK7"></a><a name="OLE_LINK6"></a><span>The concentration of</span><span> miRNA (pM)</span> | + | <a name="OLE_LINK7"></a><a |
| + | |||
| + | name="OLE_LINK6"></a><span>The concentration of</span><span> miRNA (pM)</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>C<sub>1</sub></span></i> | <i><span>C<sub>1</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The concentration of initiated probe (Abbreviated | + | <span>The concentration of initiated |
| + | |||
| + | probe (Abbreviated | ||
to iprobe)</span> | to iprobe)</span> | ||
</p> | </p> | ||
| Line 552: | Line 638: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>k<sub>1</sub></span></i> | <i><span>k<sub>1</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK8"></a><span>A constant representing | + | <a name="OLE_LINK8"></a><span>A constant |
| + | |||
| + | representing | ||
the scale factor</span> | the scale factor</span> | ||
</p> | </p> | ||
| Line 565: | Line 657: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>K<sub>m </sub></span></i> | <i><span>K<sub>m </sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>One of the characteristic constants of | + | <span>One of the characteristic |
| + | |||
| + | constants of | ||
phi29 DNA polymerase</span> | phi29 DNA polymerase</span> | ||
</p> | </p> | ||
| Line 578: | Line 676: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>V<sub>max</sub></span></i> | <i><span>V<sub>max</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>One of the characteristic constants of phi29 | + | <span>One of the characteristic |
| + | |||
| + | constants of phi29 | ||
DNA polymerase</span> | DNA polymerase</span> | ||
</p> | </p> | ||
| Line 591: | Line 695: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>k<sub>2</sub></span></i> | <i><span>k<sub>2</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span>A constant representing the scale |
| + | |||
| + | factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>V</span></i> | <i><span>V</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>The initial speed of RCA </span> | <span>The initial speed of RCA </span> | ||
| Line 615: | Line 729: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></sub></i> | + | <a |
| + | |||
| + | name="OLE_LINK9"></a><i><span>n<sub>1</sub></span></i><i><sub><span></span></sub | ||
| + | |||
| + | ></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>The moles of RCA product</span> | <span>The moles of RCA product</span> | ||
| Line 627: | Line 749: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>n<sub>2</sub></span></i> | <i><span>n<sub>2</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK10"></a><span>The number of | + | <a name="OLE_LINK10"></a><span>The |
| + | |||
| + | number of | ||
stem-loop structures in each RCA product</span> | stem-loop structures in each RCA product</span> | ||
</p> | </p> | ||
| Line 640: | Line 768: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>n</span></i> | <i><span>n</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The total amount of <a name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | + | <span>The total amount of <a |
| + | |||
| + | name="OLE_LINK23"></a><a name="OLE_LINK22"></a>stem-loop structures</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>N</span></i> | <i><span>N</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK41"></a><a name="OLE_LINK16"></a><span>The molecule number of the | + | <a name="OLE_LINK41"></a><a |
| + | |||
| + | name="OLE_LINK16"></a><span>The molecule number of the | ||
fusion protein of dCas9 and split-HRP fragments</span> | fusion protein of dCas9 and split-HRP fragments</span> | ||
</p> | </p> | ||
| Line 665: | Line 805: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>k<sub>3</sub></span></i> | <i><span>k<sub>3</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span>A constant representing the scale |
| + | |||
| + | factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>y<sub>1</sub></span></i> | <i><span>y<sub>1</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>The fluorescence intensity of | <span>The fluorescence intensity of | ||
| Line 690: | Line 840: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>N<sub>1</sub></span></i> | <i><span>N<sub>1</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of the fusion protein | + | <span>The molecule number of the fusion |
| + | |||
| + | protein | ||
of dCas9 and split-HRP fragments in the solution</span> | of dCas9 and split-HRP fragments in the solution</span> | ||
</p> | </p> | ||
| Line 703: | Line 859: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>N<sub>2</sub></span></i> | <i><span>N<sub>2</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK45"></a><span>The molecule number | + | <a name="OLE_LINK45"></a><span>The |
| + | |||
| + | molecule number | ||
of the fusion protein of dCas9 and split-HRP fragments binding with stem-loop | of the fusion protein of dCas9 and split-HRP fragments binding with stem-loop | ||
structure</span> | structure</span> | ||
| Line 717: | Line 879: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>k<sub>4</sub></span></i> | <i><span>k<sub>4</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span>A constant representing the scale |
| + | |||
| + | factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<i><span>k<sub>5</sub></span></i> | <i><span>k<sub>5</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>A constant representing the scale factor</span> | + | <span>A constant representing the scale |
| + | |||
| + | factor</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">ρ</span><span></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>Signal to noise ratio(Abbreviated to SNR)</span> | + | <span>Signal to noise ratio(Abbreviated |
| + | |||
| + | to SNR)</span> | ||
</p> | </p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">I</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of formed intact HRP | + | <span>The molecule number of formed |
| + | |||
| + | intact HRP | ||
proteins </span> | proteins </span> | ||
</p> | </p> | ||
| Line 766: | Line 956: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">I<sub>1</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The molecule number of formed intact HRP | + | <span>The molecule number of formed |
| + | |||
| + | intact HRP | ||
proteins in the solution</span> | proteins in the solution</span> | ||
</p> | </p> | ||
| Line 779: | Line 977: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">I<sub>2</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <a name="OLE_LINK43"></a><span>The molecule number | + | <a name="OLE_LINK43"></a><span>The |
| + | |||
| + | molecule number | ||
of formed intact HRP proteins through binding with stem-loop structure</span> | of formed intact HRP proteins through binding with stem-loop structure</span> | ||
</p> | </p> | ||
| Line 792: | Line 998: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">t</span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
<span>Reaction time</span> | <span>Reaction time</span> | ||
| Line 804: | Line 1,016: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| − | <td width="66" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="66" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <i><span style="font-family: | + | <i><span style="font-family:宋 |
| + | |||
| + | 体;">y<sub>2</sub></span></i> | ||
</p> | </p> | ||
</td> | </td> | ||
| − | <td width="487" valign="top" style="border:solid windowtext 1.0pt;"> | + | <td width="487" valign="top" style="border:solid |
| + | |||
| + | windowtext 1.0pt;"> | ||
<p class="MsoNormal"> | <p class="MsoNormal"> | ||
| − | <span>The signal intensity (OD<sub>450</sub>)</span> | + | <span>The signal intensity |
| + | |||
| + | (OD<sub>450</sub>)</span> | ||
</p> | </p> | ||
</td> | </td> | ||
| Line 823: | Line 1,043: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The RCA model </span></span><hr /> | + | <span><span style="color:#7f1015"> The RCA model </span></span><hr |
| + | |||
| + | /> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, we assume that there is a linear relationship between the concentration of the initiated probes and miRNA as the combination reaction between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font- |
| + | |||
| + | size:18px;">Firstly, we assume that there is a linear relationship between the | ||
| + | |||
| + | concentration of the initiated probes and miRNA as the combination reaction | ||
| + | |||
| + | between miRNA and probe occurs spontaneously<sup>1 </sup> . This can be written | ||
| + | |||
| + | as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 845: | Line 1,075: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, RCA is an enzymatic reaction, the reaction equation is written as: iprobe+ | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, |
| + | |||
| + | RCA is an enzymatic reaction, the reaction equation is written as: iprobe+dNTP→ | ||
| + | |||
| + | iprobe-dN+PPi, under the premise of excessive amount of enzymes and dNTPs, the | ||
| + | |||
| + | relationship between the concentration of iprobe and the initial speed of RCA | ||
| + | |||
| + | can be described by the Michaelis-Menten equation. This can be written as: | ||
| + | |||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 863: | Line 1,103: | ||
<!--正文--> | <!--正文--> | ||
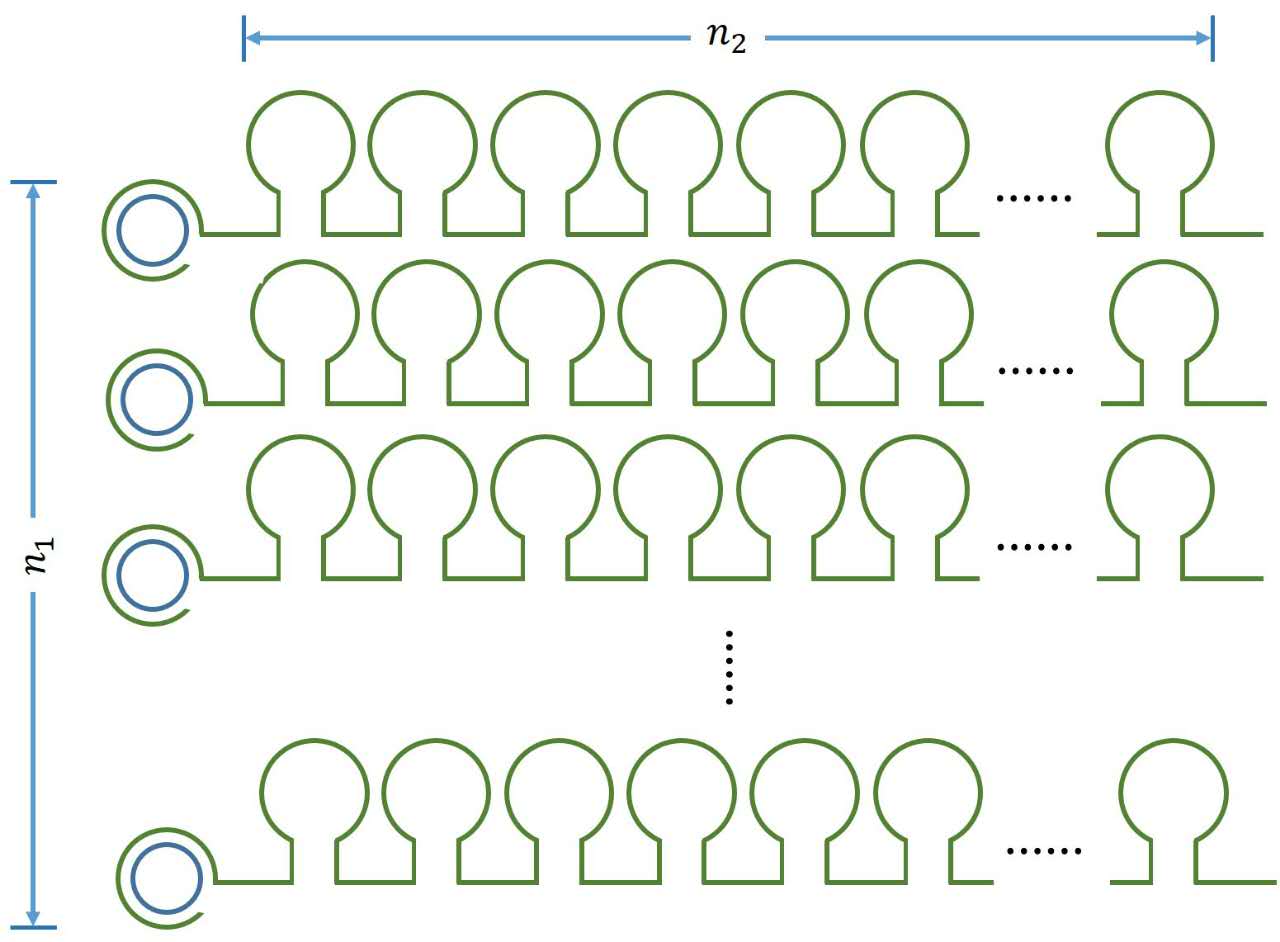
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">It is obvious that there is a linear relationship between the molecule numbers of iprobe-dN (n<sub>1 </sub> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">It is |
| − | , reflects the molecule numbers of RCA product) and the initial speed of RCA reaction. Notably, under the premise of excessive amount of enzymes, the extending of each RCA production is related to the reaction time, not the concentration of the iprobe. It is a linear relationship when we assume that the enzymatic activity remains unchanged with time, thus, the number of stem-loop structures (n<sub>2 </sub> | + | |
| − | ) in each RCA product is stable under the premise of a certain reaction time. </span> | + | obvious that there is a linear relationship between the molecule numbers of |
| + | |||
| + | iprobe-dN (n<sub>1 </sub> | ||
| + | , reflects the molecule numbers of RCA product) and the initial speed of RCA | ||
| + | |||
| + | reaction. Notably, under the premise of excessive amount of enzymes, the | ||
| + | |||
| + | extending of each RCA production is related to the reaction time, not the | ||
| + | |||
| + | concentration of the iprobe. It is a linear relationship when we assume that the | ||
| + | |||
| + | enzymatic activity remains unchanged with time, thus, the number of stem-loop | ||
| + | |||
| + | structures (n<sub>2 </sub> | ||
| + | ) in each RCA product is stable under the premise of a certain reaction time. | ||
| + | |||
| + | </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 881: | Line 1,137: | ||
<!--图注--> | <!--图注--> | ||
<p> | <p> | ||
| − | <b><span style="line-height:2;font-family:Perpetua;font-size:18px;">Figure 1. Schematic diagram</span></b> | + | <b><span style="line-height:2;font-family:Perpetua;font- |
| + | |||
| + | size:18px;">Figure 1. Schematic diagram</span></b> | ||
</p> | </p> | ||
| Line 889: | Line 1,147: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">This |
| + | |||
| + | can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 914: | Line 1,174: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And then, SYBR Green binds to DNA. The | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And |
| − | resulting DNA-dye-complex absorbs blue light (<i>λ<sub>max</sub></i> = 497nm) and emits green light (<i>λ<sub>max</sub></i> = 520nm). Thus, there is | + | |
| − | a linear relationship between the total amount of stem-loop structures and the fluorescence | + | then, SYBR Green binds to DNA. The |
| + | resulting DNA-dye-complex absorbs blue light (<i>λ<sub>max</sub></i> = 497nm) | ||
| + | |||
| + | and emits green light (<i>λ<sub>max</sub></i> = 520nm). Thus, there is | ||
| + | a linear relationship between the total amount of stem-loop structures and the | ||
| + | |||
| + | fluorescence | ||
intensity of DNA-dye-complex. This can be written as:</span> | intensity of DNA-dye-complex. This can be written as:</span> | ||
</p> | </p> | ||
| Line 931: | Line 1,197: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In summary, the relationship between the concentration of miRNA and the fluorescence intensity of DNA-dye-complex can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In |
| + | |||
| + | summary, the relationship between the concentration of miRNA and the | ||
| + | |||
| + | fluorescence intensity of DNA-dye-complex can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 939: | Line 1,209: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model6.jpg| | + | [[File:T--NUDT_CHINA--model6.jpg|210px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 947: | Line 1,217: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The signal detection model </span></span><hr /> | + | <span><span style="color:#7f1015"> The signal detection model |
| + | |||
| + | </span></span><hr /> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Firstly, it is obvious that there is a linear relationship between the molecule number of formed intact HRP proteins in the solution (consider as NOISE) and the molecule number of the fusion proteins in the solution. This can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font- |
| + | |||
| + | size:18px;">Firstly, it is obvious that there is a linear relationship between | ||
| + | |||
| + | the molecule number of formed intact HRP proteins in the solution (consider as | ||
| + | |||
| + | NOISE) and the molecule number of the fusion proteins in the solution. This can | ||
| + | |||
| + | be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 969: | Line 1,249: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, we build a probability model to find the expression of the molecule number of formed intact HRP proteins (fused with dCas9) through binding with stem-loop structure (consider as SIGNAL). It is related to the total amount of stem-loop structures and the molecule number of DNA-bound fusion proteins. We obtain the result by Monte Carlo method. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Next, |
| + | |||
| + | we build a probability model to find the expression of the molecule number of | ||
| + | |||
| + | formed intact HRP proteins (fused with dCas9) through binding with stem-loop | ||
| + | |||
| + | structure (consider as SIGNAL). It is related to the total amount of stem-loop | ||
| + | |||
| + | structures and the molecule number of DNA-bound fusion proteins. We obtain the | ||
| + | |||
| + | result by Monte Carlo method. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 984: | Line 1,274: | ||
</br> | </br> | ||
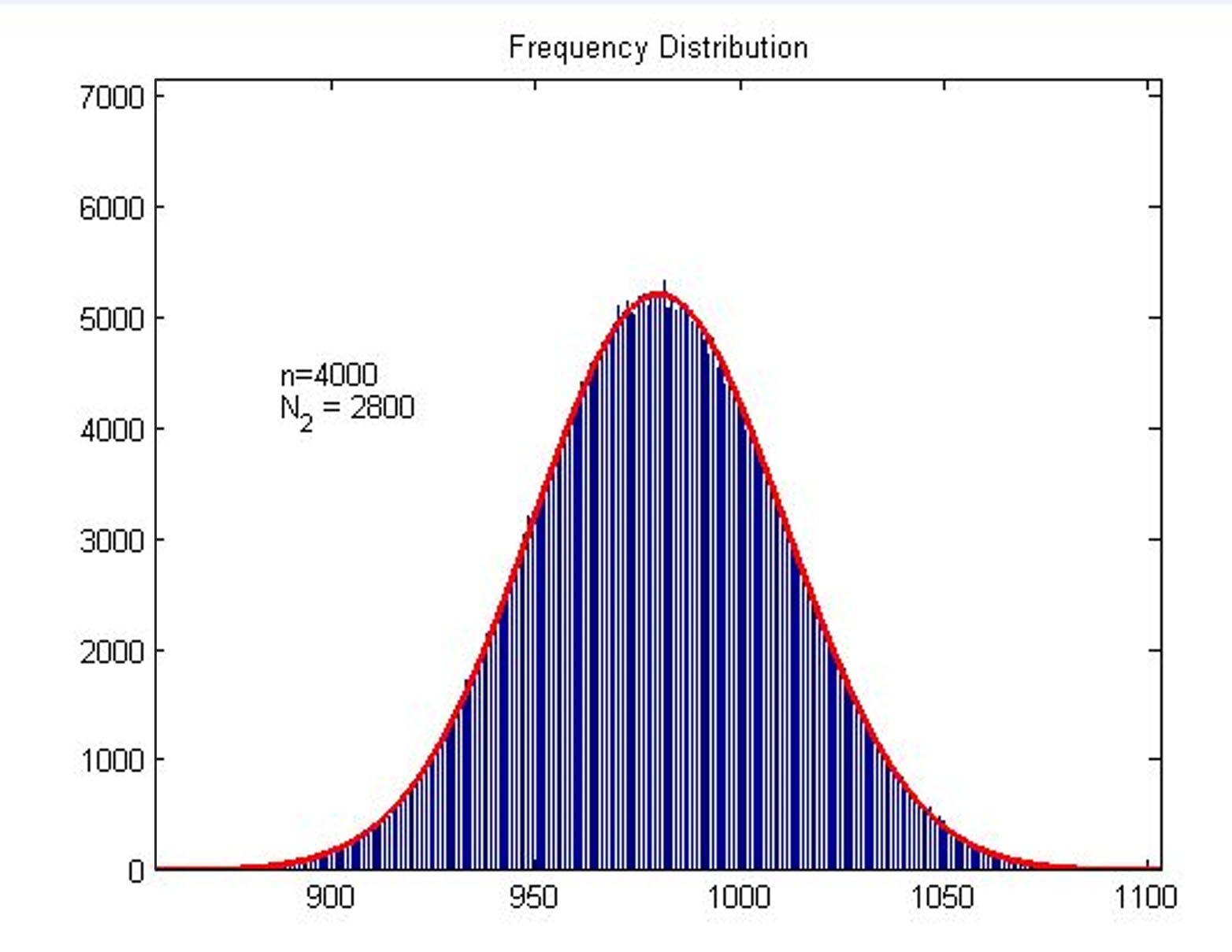
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure2. Frequency Distribution</b> Plot of frequency | ||
| + | |||
| + | distribution of the molecule number of formed intact HRP proteins through | ||
| + | |||
| + | binding with stem-loop structure. </span> | ||
</br> | </br> | ||
| Line 996: | Line 1,292: | ||
</br> | </br> | ||
<p> | <p> | ||
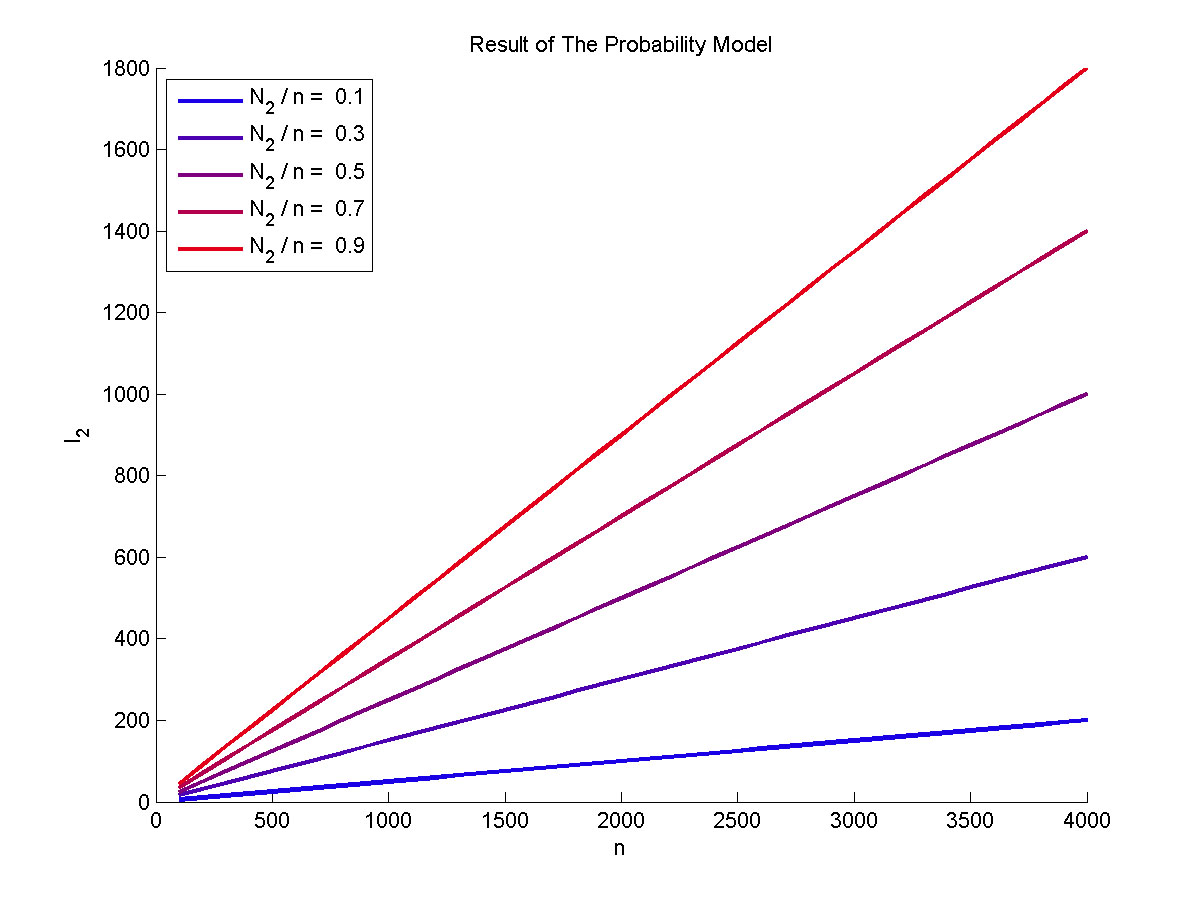
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 3. The result of the probability model</b> Plot of the | ||
| + | |||
| + | expect value of the molecule number of formed intact proteins against the total | ||
| + | |||
| + | amount of stem-loop structures, carried out at 0.1, 0.3, 0.5, 0.7, 0.9 of | ||
| + | |||
| + | N<sub>2</sub> to n ratio.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,002: | Line 1,306: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The result can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | result can be written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,011: | Line 1,317: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model8.jpg| | + | [[File:T--NUDT_CHINA--model8.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,017: | Line 1,323: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for <i>N<sub>2</sub></i>, | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">As for |
| + | |||
| + | <i>N<sub>2</sub></i>, | ||
which refers to the molecule number of the fusion proteins binding with | which refers to the molecule number of the fusion proteins binding with | ||
stem-loop structure. The equation is formulated based on the limiting case. The | stem-loop structure. The equation is formulated based on the limiting case. The | ||
| Line 1,029: | Line 1,337: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model9.jpg| | + | [[File:T--NUDT_CHINA--model9.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,036: | Line 1,344: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And then, we can obtain the expressions for <i>ρ</i></span> <span>and<i> I</i> separately.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">And |
| + | |||
| + | then, we can obtain the expressions for <i>ρ</i></span> <span>and<i> I</i> | ||
| + | |||
| + | separately.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,045: | Line 1,357: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model10.jpg| | + | [[File:T--NUDT_CHINA--model10.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,058: | Line 1,370: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Notably, under the premise of excessive substrates, | + | <span style="line-height:2;font-family:Perpetua;font- |
| + | |||
| + | size:18px;">Notably, under the premise of excessive substrates, | ||
it is a linear relationship between the reaction rate and the concentration of | it is a linear relationship between the reaction rate and the concentration of | ||
enzymes when we assume that the enzymatic activity remain unchanged with time, | enzymes when we assume that the enzymatic activity remain unchanged with time, | ||
| − | thus, we can obtain the expression of the signal intensity at <i>t<sub>th</sub> </i>time-step. This is | + | thus, we can obtain the expression of the signal intensity at <i>t<sub>th</sub> |
| + | |||
| + | </i>time-step. This is | ||
written as:</span> | written as:</span> | ||
</p> | </p> | ||
| Line 1,071: | Line 1,387: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model12.jpg| | + | [[File:T--NUDT_CHINA--model12.jpg|70px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,079: | Line 1,395: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In summary, the relationship between the number of stem-loop structures and SNR under the premise of adding a certain amount of the fusion proteins can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">In |
| + | |||
| + | summary, the relationship between the number of stem-loop structures and SNR | ||
| + | |||
| + | under the premise of adding a certain amount of the fusion proteins can be | ||
| + | |||
| + | written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,096: | Line 1,418: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The relationship between the number of Stem-loop structures and the signal intensity under the premise of adding a certain amount of the fusion proteins can be written as:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | relationship between the number of Stem-loop structures and the signal intensity | ||
| + | |||
| + | under the premise of adding a certain amount of the fusion proteins can be | ||
| + | |||
| + | written as:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,105: | Line 1,433: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model14.jpg| | + | [[File:T--NUDT_CHINA--model14.jpg|280px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,111: | Line 1,439: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> The calculation of the constants </span></span><hr /> | + | <span><span style="color:#7f1015"> The calculation of the constants |
| + | |||
| + | </span></span><hr /> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To simplify the equation (1), the constants | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">To |
| − | can be integrated. As can be seen from the above table, <i>k<sub>1</sub></i>, <i>k<sub>2</sub></i>, <i>k<sub>3</sub></i><sub> </sub>are | + | |
| − | constants representing the scale factor, <i>K<sub>m</sub></i> and <i>V<sub>max</sub></i> are | + | simplify the equation (1), the constants |
| − | characteristic constants of phi 29 DNA polymerase, and <i>n<sub>2</sub></i>, which refers to the number of stem-loop structures | + | can be integrated. As can be seen from the above table, <i>k<sub>1</sub></i>, |
| + | |||
| + | <i>k<sub>2</sub></i>, <i>k<sub>3</sub></i><sub> </sub>are | ||
| + | constants representing the scale factor, <i>K<sub>m</sub></i> and | ||
| + | |||
| + | <i>V<sub>max</sub></i> are | ||
| + | characteristic constants of phi 29 DNA polymerase, and <i>n<sub>2</sub></i>, | ||
| + | |||
| + | which refers to the number of stem-loop structures | ||
in each RCA product, is stable under the premise of a certain reaction time. | in each RCA product, is stable under the premise of a certain reaction time. | ||
| − | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation can be | + | Thus, define two constants, mark as <i>a</i> and <i>b</i>, and then the equation |
| + | |||
| + | can be | ||
simplified as:</span> | simplified as:</span> | ||
</p> | </p> | ||
| Line 1,129: | Line 1,469: | ||
</br> | </br> | ||
</html> | </html> | ||
| − | [[File:T--NUDT_CHINA--model15.jpg| | + | [[File:T--NUDT_CHINA--model15.jpg|105px|center]] |
<html> | <html> | ||
</br> | </br> | ||
| Line 1,135: | Line 1,475: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
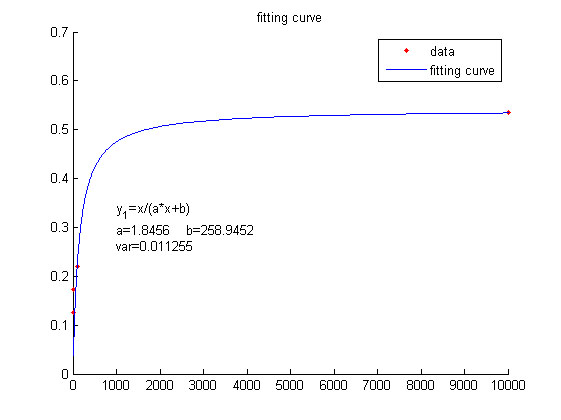
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We use this equation to fit the data points obtained through experiments. (Figure …in the ….page) The fitting curve is shown below.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">We use |
| + | |||
| + | this equation to fit the data points obtained through experiments. (Figure …in | ||
| + | |||
| + | the ….page) The fitting curve is shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,147: | Line 1,491: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 4. Fitting curve</b> Fitting curve of fluorescence | ||
| + | |||
| + | intensity of DNA-dye-complex (RFU) against the concentrations of miRNA | ||
| + | |||
| + | (pM).</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,154: | Line 1,504: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">With regard to the equation (2), we set the parameters based on the experiments as follows: | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">With |
| − | <i>k<sub>3</sub> </i>=8.35*10<sup>-16 </sup>(<i>n</i>=<i>y<sub>1</sub></i>/<i>k<sub>3</sub></i>); <i>k<sub>4</sub></i> =0.038;<i> k<sub>5</sub>t</i>=5.9*10<sup>-12</sup>. | + | |
| + | regard to the equation (2), we set the parameters based on the experiments as | ||
| + | |||
| + | follows: | ||
| + | <i>k<sub>3</sub> </i>=8.35*10<sup>-16 </sup> | ||
| + | |||
| + | (<i>n</i>=<i>y<sub>1</sub></i>/<i>k<sub>3</sub></i>); <i>k<sub>4</sub></i> | ||
| + | |||
| + | =0.038;<i> k<sub>5</sub>t</i>=5.9*10<sup>-12</sup>. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 1,163: | Line 1,521: | ||
<!--标题--> | <!--标题--> | ||
<h3> | <h3> | ||
| − | <span><span style="color:#7f1015"> Results and Analysis </span></span><hr /> | + | <span><span style="color:#7f1015"> Results and Analysis |
| + | |||
| + | </span></span><hr /> | ||
</h3> | </h3> | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By integrating the two models, we can obtain the relationships between SNR, the signal intensity respectively and the concentration of miRNA under different addition amount of fusion proteins. | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">By |
| + | |||
| + | integrating the two models, we can obtain the relationships between SNR, the | ||
| + | |||
| + | signal intensity respectively and the concentration of miRNA under different | ||
| + | |||
| + | addition amount of fusion proteins. | ||
</span> | </span> | ||
</p> | </p> | ||
| Line 1,175: | Line 1,541: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The results are shown below.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | results are shown below.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,186: | Line 1,554: | ||
</br> | </br> | ||
<p> | <p> | ||
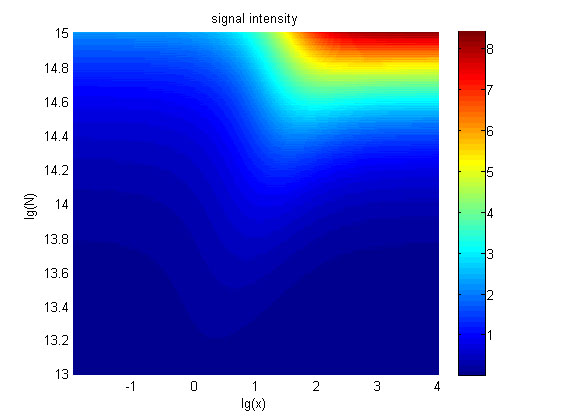
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| − | 5. The signal intensity (OD<sub>450</sub>) Three-dimensional map of signal intensity (OD<sub>450</sub>) | + | |
| − | against miRNA concentration(pM) and additional amount of fusion proteins.</span | + | size:18px;"><b>Figure |
| + | 5. The signal intensity (OD<sub>450</sub>)</b> Three-dimensional map of signal | ||
| + | |||
| + | intensity (OD<sub>450</sub>) | ||
| + | against miRNA concentration(pM) and additional amount of fusion proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,194: | Line 1,566: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">There exists a monotonous relation between the concentration of miRNA and the signal intensity when the value of the molecule number of the fusion proteins is relatively large. While the relationship does not hold when the value of the molecule number of the fusion proteins is relatively small. And the signal intensity increases as the value of the molecule number of the fusion proteins increases.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">There |
| + | |||
| + | exists a monotonous relation between the concentration of miRNA and the signal | ||
| + | |||
| + | intensity when the value of the molecule number of the fusion proteins is | ||
| + | |||
| + | relatively large. While the relationship does not hold when the value of the | ||
| + | |||
| + | molecule number of the fusion proteins is relatively small. And the signal | ||
| + | |||
| + | intensity increases as the value of the molecule number of the fusion proteins | ||
| + | |||
| + | increases.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,201: | Line 1,585: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The trend of the signal intensity with the change of the concentration of miRNA is consistent with that we obtained in our wet-lab work.(…….. in the result page)</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | trend of the signal intensity with the change of the concentration of miRNA is | ||
| + | |||
| + | consistent with that we obtained in our wet-lab work.(…….. in the result | ||
| + | |||
| + | page)</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,213: | Line 1,603: | ||
</br> | </br> | ||
<p> | <p> | ||
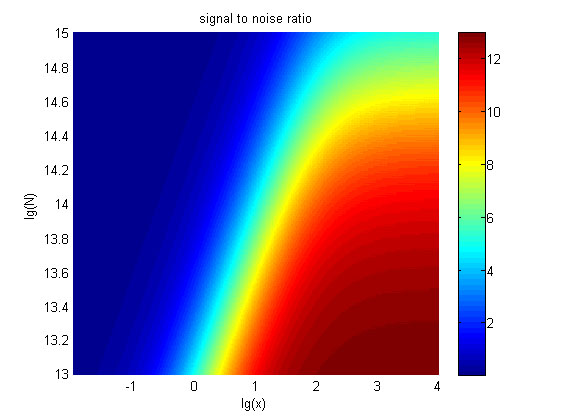
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 6. The result of SNR</b> Three-dimensional map of signal | ||
| + | |||
| + | to noise ratio against miRNA concentration (pM) and additional amount of fusion | ||
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,220: | Line 1,616: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The signal-to-noise ratio decreases as the value of the molecule number of fusion proteins increases. </span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | signal-to-noise ratio decreases as the value of the molecule number of fusion | ||
| + | |||
| + | proteins increases. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,227: | Line 1,627: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
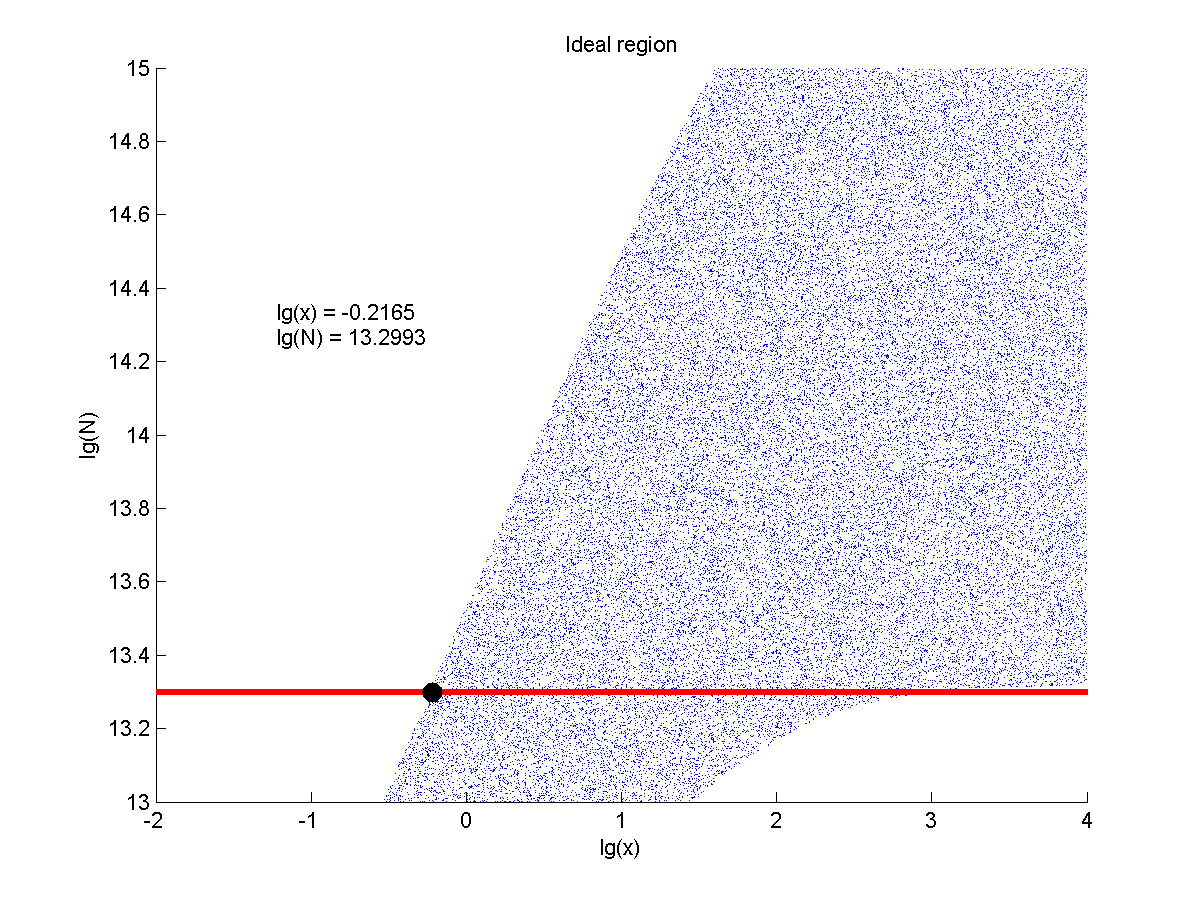
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Furthermore, we consider that it is conducive to signal detection when the values of signal intensity and signal to noise ratio are both greater than two. We obtain the ideal region through calculation, which is shown as below. </span> | + | <span style="line-height:2;font-family:Perpetua;font- |
| + | |||
| + | size:18px;">Furthermore, we consider that it is conducive to signal detection | ||
| + | |||
| + | when the values of signal intensity and signal to noise ratio are both greater | ||
| + | |||
| + | than two. We obtain the ideal region through calculation, which is shown as | ||
| + | |||
| + | below. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,240: | Line 1,648: | ||
</br> | </br> | ||
<p> | <p> | ||
| − | + | <span style="line-height:2;font-family:Perpetua;font- | |
| + | |||
| + | size:18px;"><b>Figure 7. Ideal region</b> Region of qualified logarithm of | ||
| + | |||
| + | concentrations of miRNA (pM) and logarithm of the additional amount of fusion | ||
| + | |||
| + | proteins.</span> | ||
</p> | </p> | ||
| Line 1,249: | Line 1,663: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When the value of N is set to | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">When |
| + | |||
| + | the value of N is | ||
| + | set to <a name="OLE_LINK18"></a><a name="OLE_LINK17"></a>10<sup>13.3</sup>, the | ||
| + | |||
| + | range of the value of x contained | ||
| + | in the ideal region is the largest, which means when the value of the molecule | ||
| + | number of the fusion proteins is 10<sup>13.3</sup>, the concentration range of | ||
| + | miRNA contained in the ideal region is the largest. </span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,261: | Line 1,683: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Based on our simulation, we came to the conclusion that:</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">Based |
| + | |||
| + | on our simulation, we came to the conclusion that:</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,268: | Line 1,692: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The calculated trend of the signal intensity with the change of the concentration of miRNA from our model is consistent with the trend we obtained in our laboratory work.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | calculated trend of the signal intensity with the change of the concentration of | ||
| + | |||
| + | miRNA from our model is consistent with the trend we obtained in our laboratory | ||
| + | |||
| + | work.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,274: | Line 1,704: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The value of the molecule number of fusion protein of dCas9 and split-HRP fragments not only affects the signal-to-noise ratio, but also the signal intensity. So we need to weigh its impact on both to select the optimal solution.</span> | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | value of the molecule number of fusion protein of dCas9 and split-HRP fragments | ||
| + | |||
| + | not only affects the signal-to-noise ratio, but also the signal intensity. So we | ||
| + | |||
| + | need to weigh its impact on both to select the optimal solution.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,281: | Line 1,717: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The concentration range of miRNA contained in the ideal region is the largest when we set the value of the molecule number of fusion proteins to be | + | <span style="line-height:2;font-family:Perpetua;font-size:18px;">The |
| + | |||
| + | concentration range | ||
| + | of miRNA contained in the ideal region is the largest when we set the value of | ||
| + | the molecule number of the fusion proteins to be 10<sup>13.3</sup>, we optimize | ||
| + | the value of the addition amount of fusion proteins in our wet-lab work based | ||
| + | on it.</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| Line 1,293: | Line 1,735: | ||
<!--正文--> | <!--正文--> | ||
<p style="text-indent:22pt;"> | <p style="text-indent:22pt;"> | ||
| − | <span style="line-height:2;font-family:Perpetua;font-size:18px;">1 Deng, R. <i>et al</i>. Toehold-initiated rolling circle | + | <span style="line-height:2;font-family:Perpetua;font- |
| − | amplification for visualizing individual microRNAs in situ in single cells. <i>Angew Chem Int Ed Engl </i>53, 2389-2393, | + | |
| + | size:18px;">1 Deng, R. <i>et al</i>. | ||
| + | |||
| + | Toehold-initiated rolling circle | ||
| + | amplification for visualizing individual microRNAs in situ in single cells. | ||
| + | |||
| + | <i>Angew Chem Int Ed Engl </i>53, 2389-2393, | ||
doi:10.1002/anie.201309388 (2014).</span> | doi:10.1002/anie.201309388 (2014).</span> | ||
</p> | </p> | ||
</br> | </br> | ||
| + | |||
| + | |||
Revision as of 01:34, 17 October 2016