| Line 270: | Line 270: | ||
<div class="container"> | <div class="container"> | ||
| − | <h3 class="title1"><span> | + | <h3 class="title1">3.<span>QUESTIONNAIRE</span> COLLABORATION</h3> |
<p style="font-size:20px">We team Tianjin contacted <a href="https://2016.igem.org/Team:Pasteur_Paris">Team Pasteur</a>, <a href="https://2016.igem.org/Team:Pittsburgh">Team Pittsburgh</a>, <a href="https://2016.igem.org/Team:Valencia_UPV">Team Valencia UPV</a>, <a href="https://2016.igem.org/Team:Virginia">Team Virginia</a> and <a href="https://2016.igem.org/Team:NTU-Singapore">Team NTU</a> about the collaboration in July. Then we established and led a survey union with the support of them. We asked each team submit two or three questions about their own project and one question about synthetic biology or iGEM, and integrated the questions to the final version questionnaire.<br/> </br> | <p style="font-size:20px">We team Tianjin contacted <a href="https://2016.igem.org/Team:Pasteur_Paris">Team Pasteur</a>, <a href="https://2016.igem.org/Team:Pittsburgh">Team Pittsburgh</a>, <a href="https://2016.igem.org/Team:Valencia_UPV">Team Valencia UPV</a>, <a href="https://2016.igem.org/Team:Virginia">Team Virginia</a> and <a href="https://2016.igem.org/Team:NTU-Singapore">Team NTU</a> about the collaboration in July. Then we established and led a survey union with the support of them. We asked each team submit two or three questions about their own project and one question about synthetic biology or iGEM, and integrated the questions to the final version questionnaire.<br/> </br> | ||
Then, we posted it on twitter and facebook, many iGEMers participated in it and many iGEM teams reposted it on their homepage. We are very delighted to see their enthusiasm which encourages us a lot!</p> | Then, we posted it on twitter and facebook, many iGEMers participated in it and many iGEM teams reposted it on their homepage. We are very delighted to see their enthusiasm which encourages us a lot!</p> | ||
| Line 287: | Line 287: | ||
<div class="gallery"> | <div class="gallery"> | ||
<div class="container"> | <div class="container"> | ||
| − | <h3 class="title1">4. | + | <h3 class="title1">4.COMMUNICATION <span>WITH </span><a href="https://2016.igem.org/Team:Peking">PKU</a></h3> |
<p style="font-size:20px"> Six representatives of Tianjin University iGEM 2016 Team arrived Peiking University on a rainy afternoon of 19th August and had a long lovely dicussion meeting with three other teams, Beijing Institute of Technology(Team BIT-China),University of Chinese Academy of Science(Team UCAS) and the ebullient host, Peiking University iGEM team.</br></br> | <p style="font-size:20px"> Six representatives of Tianjin University iGEM 2016 Team arrived Peiking University on a rainy afternoon of 19th August and had a long lovely dicussion meeting with three other teams, Beijing Institute of Technology(Team BIT-China),University of Chinese Academy of Science(Team UCAS) and the ebullient host, Peiking University iGEM team.</br></br> | ||
On the very beginning of the meeting, each team leader gave a brief introduction on their projects. And team Tianjin's project attracted great interests in the environment problem of PET-degradation. After each team's presentation, we started to share our new ideas and problems we crossed with during the experiment, which was a very helpful and encouraging session where we help each other fix the experimental problems.</br></br> | On the very beginning of the meeting, each team leader gave a brief introduction on their projects. And team Tianjin's project attracted great interests in the environment problem of PET-degradation. After each team's presentation, we started to share our new ideas and problems we crossed with during the experiment, which was a very helpful and encouraging session where we help each other fix the experimental problems.</br></br> | ||
| Line 382: | Line 382: | ||
<div class="container"> | <div class="container"> | ||
<span class="bg-text" style="z-index:-9999">Tianjin</span> | <span class="bg-text" style="z-index:-9999">Tianjin</span> | ||
| − | <h3 class="title1">6.OUR HP <span> | + | <h3 class="title1">6.OUR HP <span>WITH</span> <a href="https://2016.igem.org/Team:Nanjing-China">TEAM NJU iGEM</a></h3> |
<div class="testi-info"> | <div class="testi-info"> | ||
| Line 528: | Line 528: | ||
<div class="welcome about"> | <div class="welcome about"> | ||
<div class="container"> | <div class="container"> | ||
| − | <h3 class="title1"> | + | <h3 class="title1">7.OUR COLLABORATION <span>WITH </span> <a href="https://2016.igem.org/Team:Jilin_China">Jilin_China</a> </h3> |
</br> | </br> | ||
<div class="col-md-3 about-right wow fadeInRight animated" data-wow-delay=".5s"> | <div class="col-md-3 about-right wow fadeInRight animated" data-wow-delay=".5s"> | ||
<figure> | <figure> | ||
| − | <a href="https://static.igem.org/mediawiki/2016/ | + | <a href="https://static.igem.org/mediawiki/2016/c/c4/T--Tianjin--jilin.png" data-lightbox="no" data-title="Discovery Studio 3.5"> |
| − | <img src="https://static.igem.org/mediawiki/2016/ | + | <img src="https://static.igem.org/mediawiki/2016/c/c4/T--Tianjin--jilin.png" style="width:120%"> |
</a> | </a> | ||
| − | <figcaption> | + | <figcaption> Discovery Studio 3.5 </figcaption> |
</figure> | </figure> | ||
| Line 542: | Line 542: | ||
<div class="col-md-9 about-left wow fadeInLeft animated" data-wow-delay=".5s"> | <div class="col-md-9 about-left wow fadeInLeft animated" data-wow-delay=".5s"> | ||
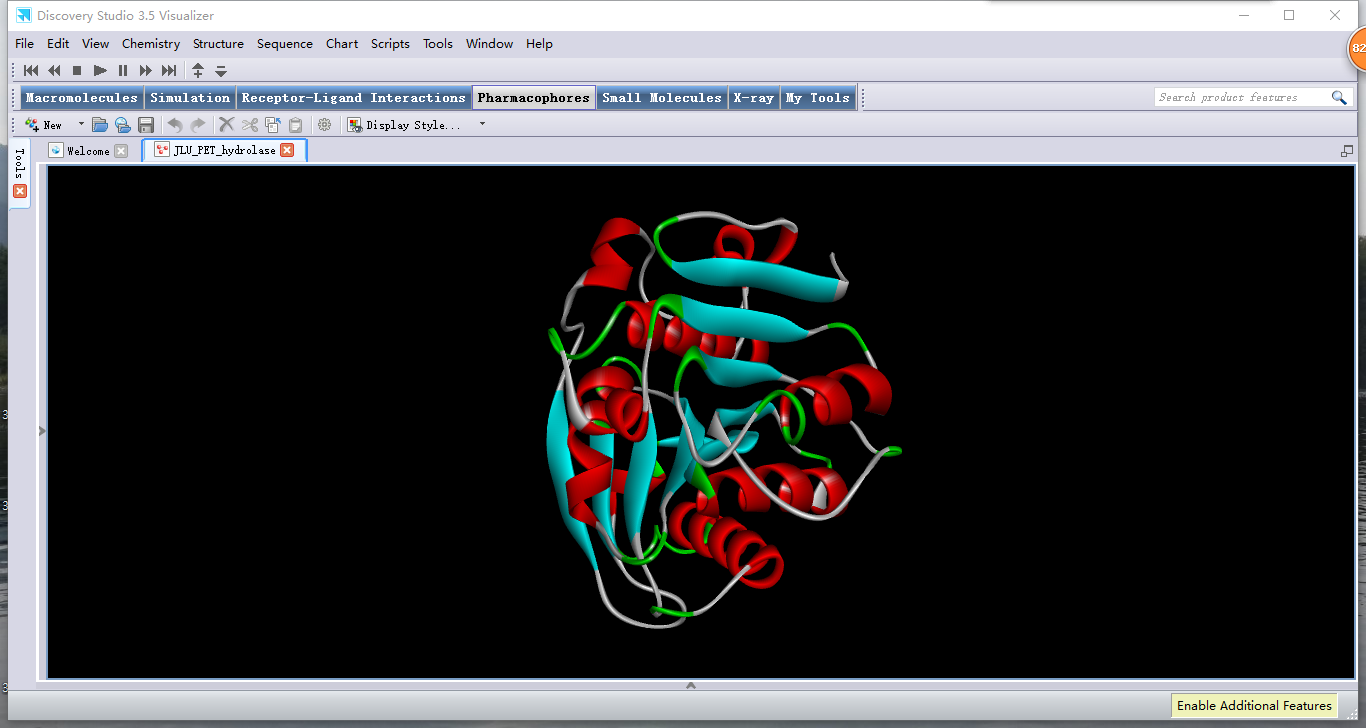
| − | <p class="data" style="color:#344525;font-size:20px"> | + | <p class="data" style="color:#344525;font-size:20px">In the process of designing the site-directed mutation, we communicate with Team Jilin_China about our problem. They offered us a software Discovery Studio 3.5, which can speculate the rough 3D structure of a protein molecule according to its amino acid sequence. This software gave us a reference about which site should be mutated without the exact 3D structure. |
| − | + | ||
| − | + | ||
<br /> | <br /> | ||
</div> | </div> | ||
| Line 559: | Line 557: | ||
<div class="container"> | <div class="container"> | ||
<span class="bg-text" style="z-index:-9999">Tianjin</span> | <span class="bg-text" style="z-index:-9999">Tianjin</span> | ||
| − | <h3 class="title1"> | + | <h3 class="title1">8.SYSU-CCiC <span>MEETING</span></h3> |
<div class="testi-info"> | <div class="testi-info"> | ||
Revision as of 14:41, 18 October 2016
<!DOCTYPE html>
1.OUR HP With TEAM HFUT iGEM
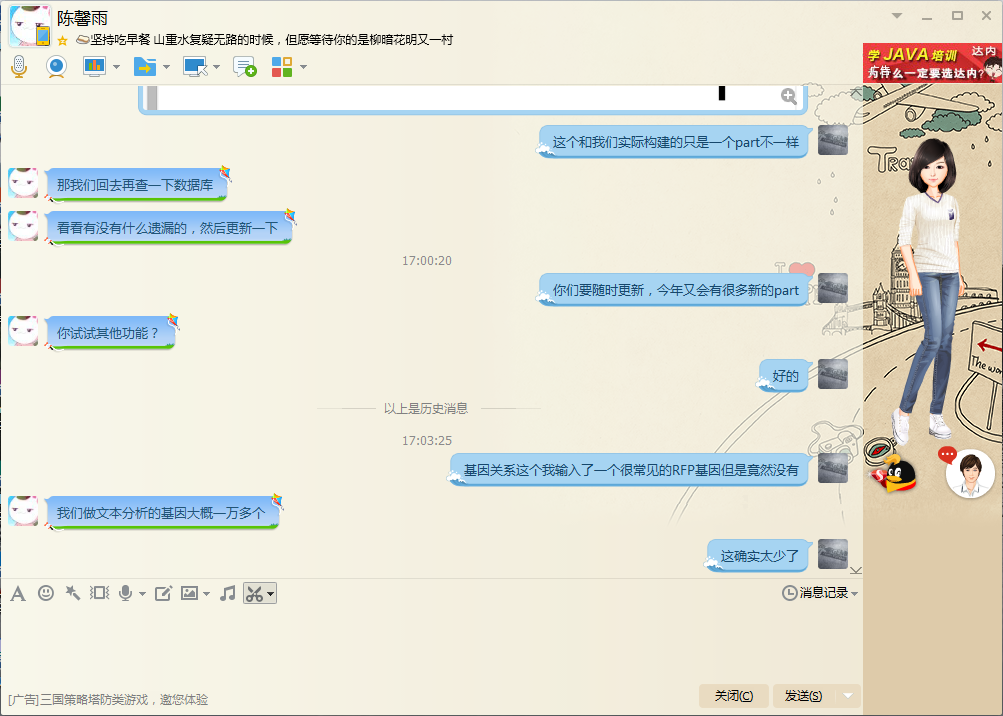
In this section of collaboration, we helped the Team HFUT test their software Biodesigner Coral, an online software that can help users search for particular part of iGEM and construct their own part, the relationship among genes, compounds, or parts, and offer the detailed information of a plenty of genes. We discussed about the software according to the using experience of our team. We tried to use this software to build a part we use this year and search for some genes we often use. We gave them some suggestions online. Here are the screenshot pictures of the online chatting records. (We use the common social network QQ to communicate, Haoran Ding represented our team and Xinyu Chen reprensented Team HFUT)
Ding: What is more, Your database may be incomplete, for I searched for the part BBa_E1010 used in one of our device, but I did not get the result even if I directly type the part number. Ding: (screenshot, to show the lack of result when search for a particular part)
Ding: And there is a BBa_E1011, but it is codon optimized mRFP-1 gene in yeast. Ding: But I do not need this optimization. Ding: (screenshot, to show the information on the box on the right of the page) Ding: Actually I have found this part in the information box on the right of the page. Ding: Of course it is excellent overall, I have constructed a device according to the guide. Ding: (screenshot, to show the finished device) Ding: This device has only one part different from the actually constructed part.
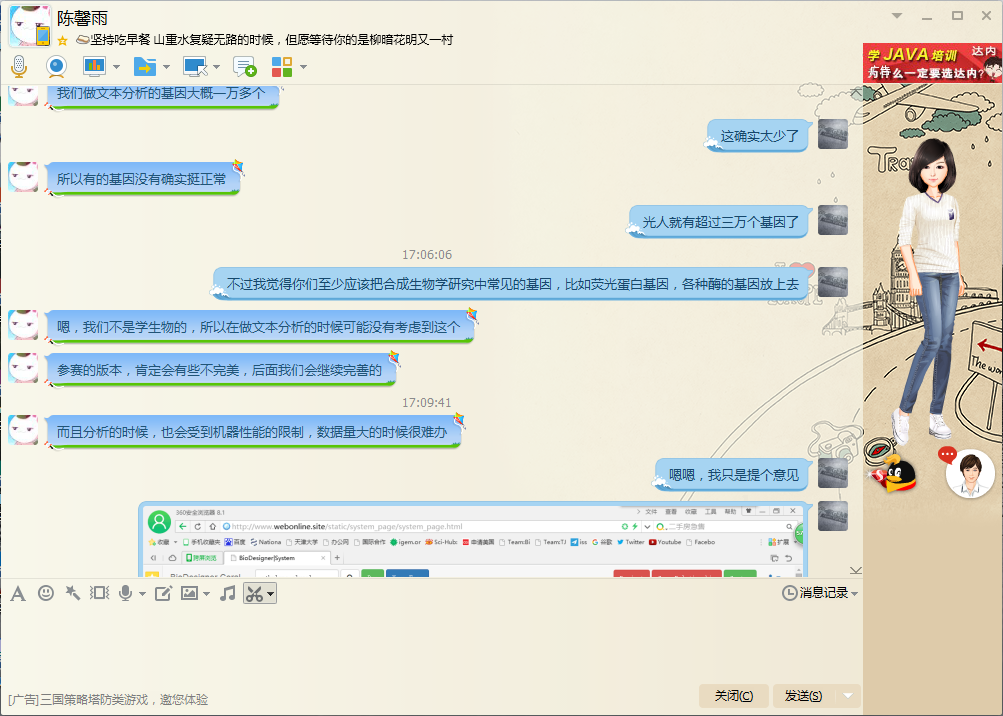
Chen: I will check the database after a while. Chen: To see if there is something missed and update the database. Ding: You need to update at all time, this year many new parts will enter the iGEM part database. Chen: Would you like try some other functions? Ding: OK. Ding: I enter a common gene RFP but I unexpectedly get no result. Ding: It is really a problem.
Chen: Thank you for your remind. You know there are so many genes in the world, Restricted by time and finance, and even the quality of machine, we can not analysis all genes in the world. Ding: I can understand it. It is hard to integrate so many genes' data. Ding: But I do think that you should upload the common genes in the synthetic biology researching process like genes of fluorescent protein, different kinds of enzymes and so on. Chen: OK, because we are not major in biology, so we did not think about this when we did the text analysis. Chen: It is unavoidably imperfect for this edition. We will perfect it in the future. Ding: Yeah, it is just a suggestion.
Ding: I have not found any problems in the System page. Ding: In summary, this software is excellent, I guess it is the first software focusing on the searching of iGEM part. Ding: What you need to do next is enlarge the database, remove some bugs like unable to load picture, and change the part editing page, which I think is not so friendly. Chen: OK, thank you for your feedback. Ding: You are welcome.
2.OUR LECTURE IN TUST
Tianjin University of Science and Technology is a key university in Tianjin, China, which is famous for its excellent fermentation engineering and food engineering subject. Some years ago, some professors and students of TUST decided to set up a iGEM team and participate in the iGEM competition in the future. This summer, a sophomore student and a graduate student from TUST came to Tianjin University and study the basic knowledge about iGEM and synthetic biology and help our experiments in protein modification part. They told us about the enormous difficulty they are facing in team establishing process and we shared our experience and gave several suggestions to them. Before they leaved us, they invited us to give a speech in their school after summer vacation. On September 27th, five members of our team went to Teda campus of TUST and gave a speech about how to set up a iGEM team and what could a iGEMer receive from this competition. Almost all the seats were occupied by listeners. Since the iGEM competition is almost a band new term for most students in TUST, many of them showed great interests and talk with us enthusiastically. We gave them a plenty of suggestions according to the experience of our project, and offered encouragement to the students there. We continued our communication online by using a popular Chinese social network Wechat. They will recruit members this semester and decided to register a team next year.
3.QUESTIONNAIRE COLLABORATION
We team Tianjin contacted Team Pasteur, Team Pittsburgh, Team Valencia UPV, Team Virginia and Team NTU about the collaboration in July. Then we established and led a survey union with the support of them. We asked each team submit two or three questions about their own project and one question about synthetic biology or iGEM, and integrated the questions to the final version questionnaire.
Then, we posted it on twitter and facebook, many iGEMers participated in it and many iGEM teams reposted it on their homepage. We are very delighted to see their enthusiasm which encourages us a lot!

4.COMMUNICATION WITH PKU
Six representatives of Tianjin University iGEM 2016 Team arrived Peiking University on a rainy afternoon of 19th August and had a long lovely dicussion meeting with three other teams, Beijing Institute of Technology(Team BIT-China),University of Chinese Academy of Science(Team UCAS) and the ebullient host, Peiking University iGEM team.
On the very beginning of the meeting, each team leader gave a brief introduction on their projects. And team Tianjin's project attracted great interests in the environment problem of PET-degradation. After each team's presentation, we started to share our new ideas and problems we crossed with during the experiment, which was a very helpful and encouraging session where we help each other fix the experimental problems.
In a nutshell, this short trip to Beijing is a win-win collaboration.
5.OUR HP WITH TEAM NKU iGEM
On the day we visited professor Cunjiang Song, we conduct a human practice with team Nankai iGEM.
We visited team Nankai’s laboratory in Nankai university. They introduced to us about the general situation of them. We discussed with their members about project affairs about life in the laboratory. We took a photo together finally.
We are delighted to communicate with members of NKU iGEM. We hope that our rapport and friendship would last forever!
6.OUR HP WITH TEAM NJU iGEM
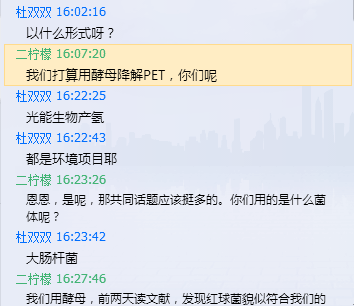
We found that Rhodococcus can degrade intermediate product TPA efficiently. Thus we got in touch with Prof. Aijun Liao with the help of iGEM NJU. He presented us the Rhodococcus jostii RHA1 which helps us a lot. We are grateful to what he has done.
Du:
Hi!
Xu:
Hi!
Du:
I’m Shuangshuang Du of team Nanjing_China from Nanjing University.
Xu:
I’m a member of iGEM_Tianjin. Our team would like to communicate with you.
Du:
Sure.
Du:
By what means?
Xu:
We intend to biodegrade PET using yeasts. How about your project?
Du:
Photobiological hydrogen production.
We both are environmental projects!
Xu:
Yes. Then we should have plenty of common topics. What strains do you use?
Du:
We use Escherichia coli.
Xu:
We use yeasts, but the other day when we consulted theses, we found that Rhodococcus might meet some of our needs.
We use yeasts, but the other day when we consulted theses, we found that Rhodococcus might meet some of our needs. Searching through the internet, we learnt that Pro. Wanjun Liao from your university had deep researches in this area, so I wonder if you are familiar with him?
Du:
No. Though I’m not acquainted with him, I can ask my schoolmate in the School of Environment about him for you.
Xu:
Oh, yes. We’d really appreciate your help! By the way, do you have interflow with Zhejiang University at the moment?
Du:
I took part in its summer courses personally, and the courses have ended.
We don’t conduct any interflow with Zhejiang University currently.
Xu:
I see. We were engaged in our final exams some time ago and just began our project, too.
Du:
Hello!
The teacher replied and agreed.
Please send your address to me.
Xu:
Ha-ha!
Du:
So I think I’ll post it to you tomorrow by SF express.
Xu:
Lots of thanks! I’ll give you our address right now.
7.OUR COLLABORATION WITH Jilin_China
In the process of designing the site-directed mutation, we communicate with Team Jilin_China about our problem. They offered us a software Discovery Studio 3.5, which can speculate the rough 3D structure of a protein molecule according to its amino acid sequence. This software gave us a reference about which site should be mutated without the exact 3D structure.
8.SYSU-CCiC MEETING
It was a great honor to be invited to SYSU-CCiC meeting organized by Sun Yat-Sen University (SYSU) to share iGEM experiences with other teams. CCiC(Central China iGEM Consortium) is a national iGEM seminar held by Chinese universities, aiming to promote the communication of Chinese iGEM teams and publicize the conception of synthesis biology.
On the morning of September 2, 2016, we took a flight to Guangzhou from Tianjin. One hour later, we arrived at SYSU. Upon our registration, we exchanged ideas with some teams. At 7 pm, the opening ceremony began and each team made a brief introduction. In the following two days, we simulated the process of iGEM Giant Jamboree, presenting team projects and posters.
At 5 pm of September 4,2016, the seminar was coming to a close. We took a photo together as a reminder. After dinner, we attended the closing ceremony. Afterwards, we went back to Tianjin and got occupied with our experiment again.
We are so grateful to SYSU for invitation and treatment. During the seminar, we have made a lot of friends and had an understanding of other teams’ projects. In the meanwhile, we also got a number of beneficial suggestions.