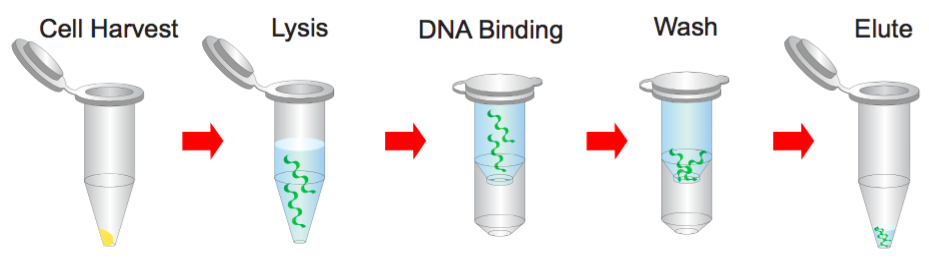
Step 1 Bacterial Cells Harvesting
1. Transfer 1.5 ml bacterial culture to a microcentrifuge tube.
2. Centrifuge at 14,000 x g for 1 minute and discard the supernatant.
Step 2 Resuspend
1. Resuspend pelleted bacterial cells in 200 μl of the Buffer S1 (RNase A added).
Step 3 Lysis
1. Add 200 μl of the Buffer S2 and mix thoroughly by inverting the tube 10 times (do not vortex) and then stand at the room temperature for 2 minutes or until the lysate is homologous.
Step 4 Neutralization
1. Add 300 μl of the Buffer S3 and mix immediately and thoroughly by inverting the tube 10 times (Do not vortex).
2. Centrifuge at 14,000 x g for 3 minutes.
Step 5 Binding
1. Place a PM column in a Collection Tube. Apply the supernatant (from step 4) to the PM column by decanting or pipetting.
2. Centrifuge at 14,000 x g for 30 seconds, then discard the flow-through, and place the PM column back into the same col/lection tube.
Step 6 Wash
1. Add 400 μl of the Buffer W1 into the PM column.
2. Centrifuge at 14,000 x g for 30 seconds.
3. Discard the flow-through and place the PM column back into the same collection tube.
4. Add 600 μl of the Buffer W2 (Ethanol added) into the PM column.
5. Centrifuge at 14,000 x g for 30 seconds.
6. Discard the flow-through and place the PM column back into the same collection tube.
7. Centrifuge at 14,000 x g again for 2 minutes to remove the residual Buffer W2.
Step 7 Elution
1. To elute DNA, place the PM column in a clean 1.5 ml microcentrifuge tube.
Add 50~200 μl of the Buffer E or H2O (pH between 7.0 and 8.5) to the center of each PM column, let it stand for 2 minutes, and centrifuge at 14,000 x g for 2 minutes.
NOTE: Check the buffers before use for salt precipitation. Redissolve any precipitate by warming to 37°C.
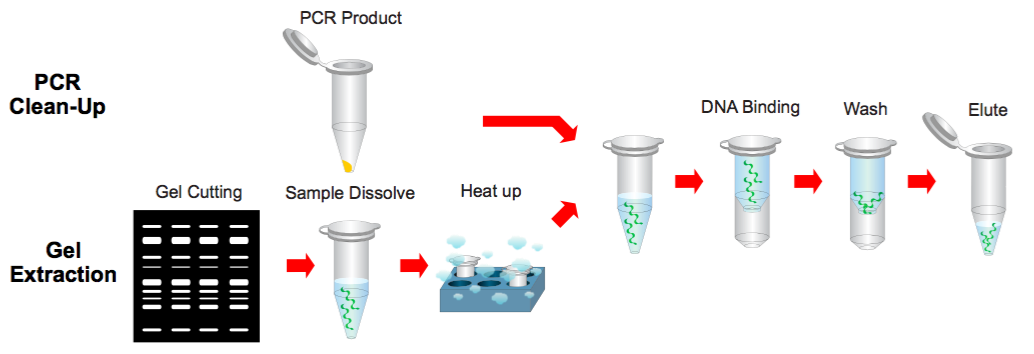
Step 1 Sample Preparation
PCR Clean Up
1. Add 500 μl of the Buffer B to 100 μl of the PCR product and mix by vortex.
Gel Extraction
1. Excise the DNA fragment from the agarose gel.
2. Transfer up to 300 mg of the gel slice to a 1.5 ml microcentrifuge tube.
3. Add 500 μl of the Buffer B to the sample and mix by vortex. Incubate at 60°C for 10 minutes (or until the gel slice has completely dissolved).
4. During the incubation, mix by vortexing the tube every 2~3 minutes.
5. Cool the dissolved sample mixture to the room temperature.
Step 2 Binding
1. Place a PG Column in a Collection Tube. Apply the supernatant (from step 1) to the PG Column by decanting or pipetting.
2. Centrifuge at 14,000 x g for 30 seconds.
3. Discard the flow-through and place the PG Column back into the same collection tube.
* The maximum volume of the PG Column reservoir is 800 μl. If the sample mixture is more than 800 μl, repeat the DNA Binding Step.
Step 3 Wash
1. Add 400 μl of the Buffer W1 into the PG Column.
2. Centrifuge at 14,000 x g for 30 seconds.
3. Discard the flow-through and place the PG Column back into the same collection tube. 4. Add 600 μl of the Buffer W2 (ethanol added) into the PG Column.
5. Centrifuge at 14,000 x g for 30 seconds.
6. Discard the flow-through and place the PG Column back into the same collection tube.
7. Centrifuge at 14,000 x g again for 2 minutes to remove the residual Buffer W2.
Step 4 Elution
1. To elute the DNA, place the PG Column in a clean 1.5 ml microcentrifuge tube.
2. Add 50-200 μl of the Buffer E or H2O (pH is between 7.0 and 8.5) to the center of each PG Column, let it stand for 2 minutes, and centrifuge at 14,000 x g for 2 min.
NOTE: Check the buffers before use for salt precipitation. Redissolve any precipitate by warming to 37°C.