Experiments
When it comes down to it, SMITe consists of three main components: a spider silk scaffold provided by our sponsors, biofilm-degrading combat proteins and the Sortase A enzyme used to conjugate the one to the other. Our main goals were the expression of functional Sortase A and combat proteins as well as their conjugation to the silk scaffold. Below, you can find an overview of the paths we chose - you can also have a look at our protocols, results or lab books if you’re interested in more details.
Combat Proteins
All of our biofilm-degrading combat proteins were assembled in a similar fashion, allowing for the addition of a linker tag sequence (LT) that would include sequences for both purification of the proteins via Immobilized Metal Affinity Chromatography (IMAC) and their conjugation to the spider silk using Sortase A.
Assembling our Constructs
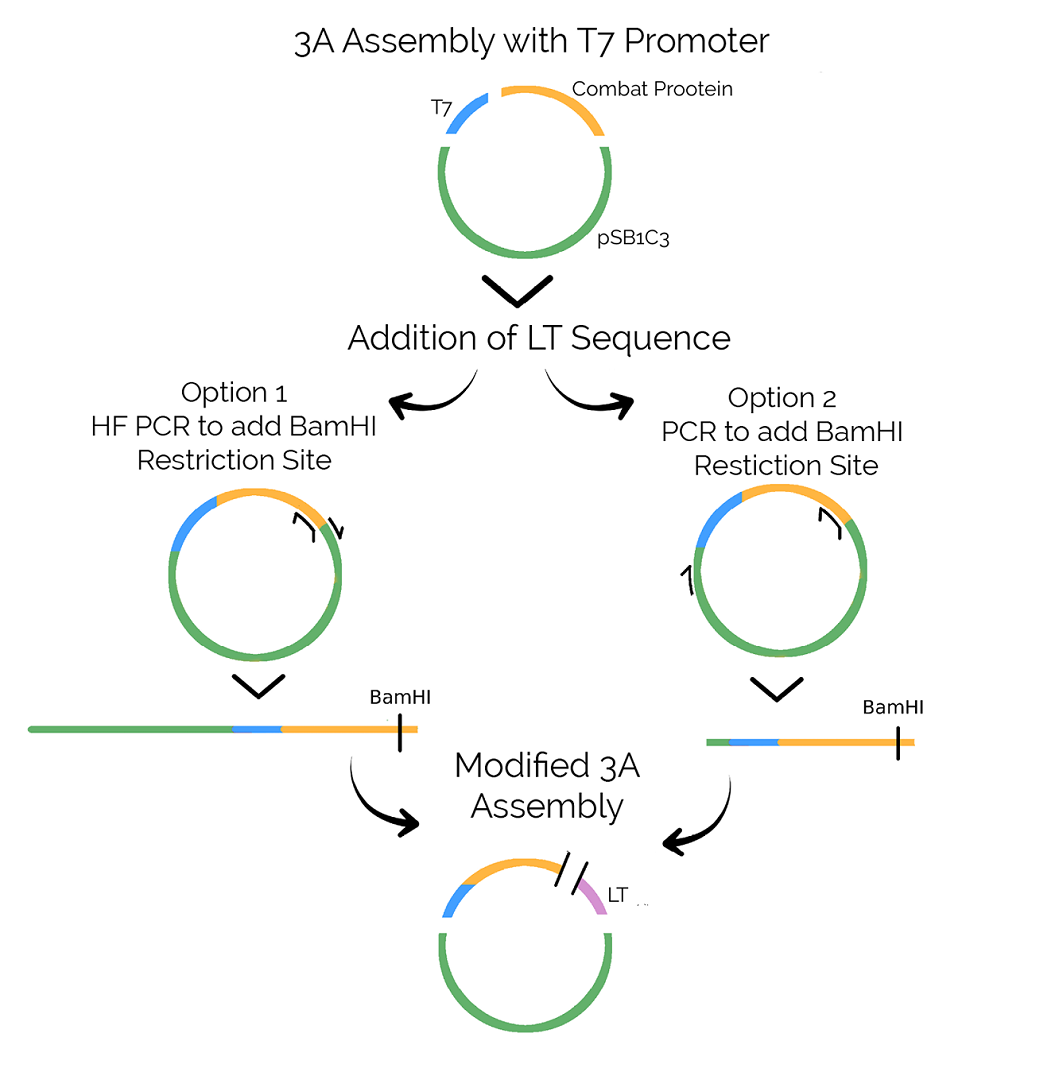
Assembly was conducted in two separate steps. First, we used the classic 3A method to assemble our Combat Proteins - defensin, lysostaphin, Nuc and esp - with a T7 promoter. Subsequently, we tested two methods to add the linker tag sequence that was needed for purification and conjugation to the spider silk.
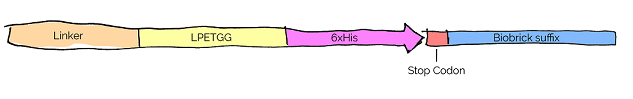
The custom designed LT sequence consists of three elements: a GSSSS linker, a Hisx6 tag for IMAC purification and LPETGG, the C-terminal sequence motif recognized by Sortase A.
Both options rely on a PCR reaction to add a BamHI restriction site to the N-terminus of the combat proteins so that BamHI can be used to ligate the T7-Combat Protein to the LT oligonucleotide. A modified 3A Assembly was used to ligate the T7-Combat Protein fragment with the LT and the pSB1C3 backbone.
Testing Combat proteins
Once our combat proteins were assembled with the promoters, we needed to test whether they were expressed correctly and decided to use a simple SDS-PAGE for that purpose. Subsequently, we tested the combat proteins' activity, purified them and attached them to spider silk.
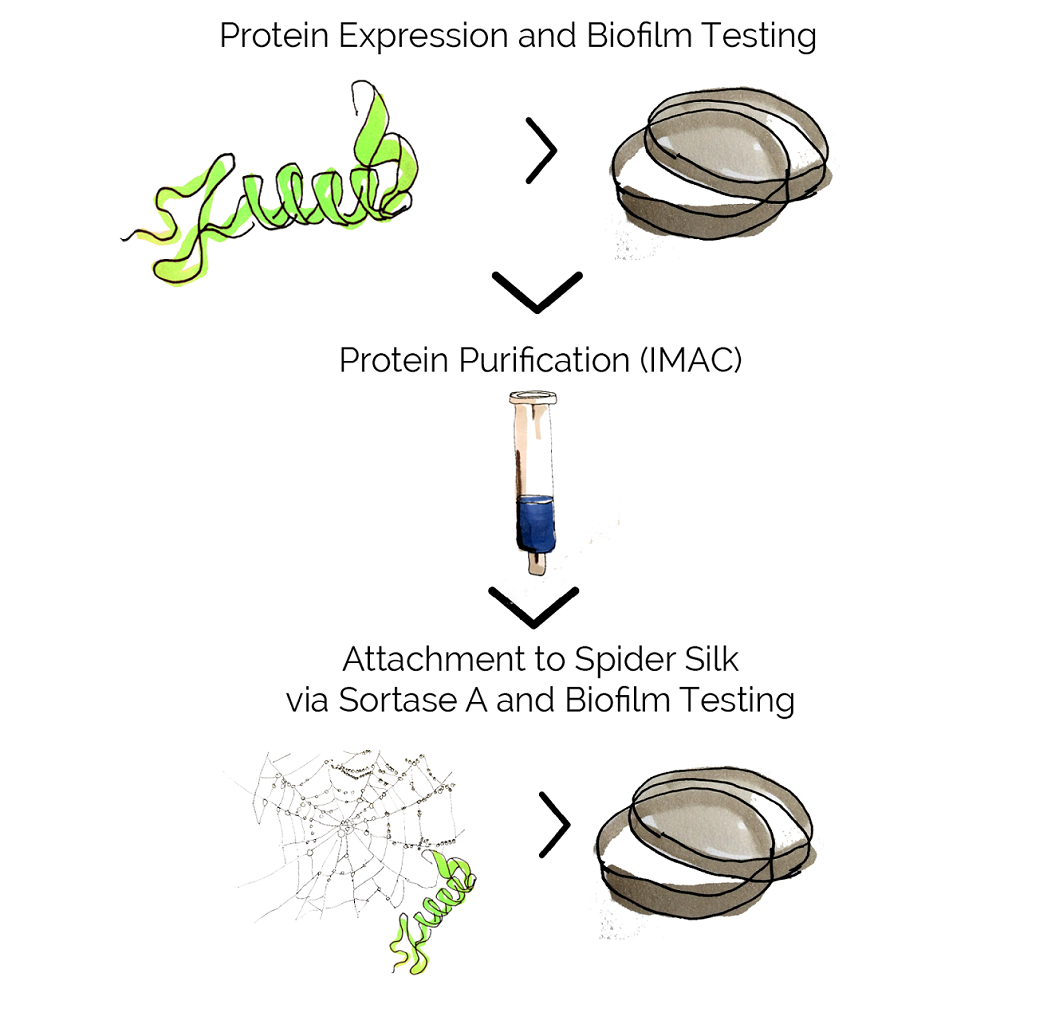
To test the functionality of the combat proteins, we decided to work primarily with biofilm assays using S. aureus and P. aeruginosa since they provide a quantifiable, clinically relevant readout. Simply put, the bacteria are incubated in conditions allowing them to form a biofilm and then the biofilm-degrading proteins are added either straight away (providing data regarding the inhibition of biofilm formation) or after biofilm had been established (measuring the ability to disperse biofilm). At different timepoints, the biofilm was stained, liquified and the absorption measured in a spectrophotometer.
Apart from the biofilm assay, we also used modified Kirby-Bauer tests where filter papers soaked in cell lysates were placed onto agar plates with bacterial cultures streaked on them. If the enzymes within the cell lysates were bacteriolytic or bacteriostatic, they inhibited the colony formation in their environment, leading to halo formation around the filter papers.
Conjugating Combat Proteins to Spider Silk
Once the combat proteins were assembled with an appropriate LT sequence and purified, the conjugation was going to be fairly straightforward. Sortase A is able to catalyze the formation of a peptide bond in between two glycines - one found on the LPETGG sequence of the combat proteins' tag and the other one naturally present on the spider silk.
Once the wound dressing components are fully assembled, more biofilm tests could be performed, alongside different stainings to visualize how the components of the biofilm are broken down in response to treatment.
Sortase A
Sortase A is our matchmaker enzyme and is used to conjugate combat proteins to the spider silk. Its sequence was easily isolated from S. aureus genome and subsequently, two modifications from the wild type Sortase A were planned to improve solubility:
- Truncation of the enzymes so that the transmembrane domain (amino acids 1-59) would not be included in our BioBrick
- N-terminal addition of GB1, a Protein G domain well known to increase solubility of recombinant proteins
Additionally, an endogenous XbaI site was aimed to be removed via site-directed mutagenesis in order to comply with BioBrick standards.
Assembling the Construct
Sortase A was isolated from S. aureus genomic DNA using a simple PCR reaction. In order to add the GB1 domain, two methods were used in the lab - a Gibson assembly and an Overlap Extension PCR. Both of those methods require homologous sequences on the fragments that will be ligated. Those overlaps were added to Sortase A with another PCR reaction, which also allowed the introduction of a C-terminal Histidine tag for IMAC purification of Sortase A.
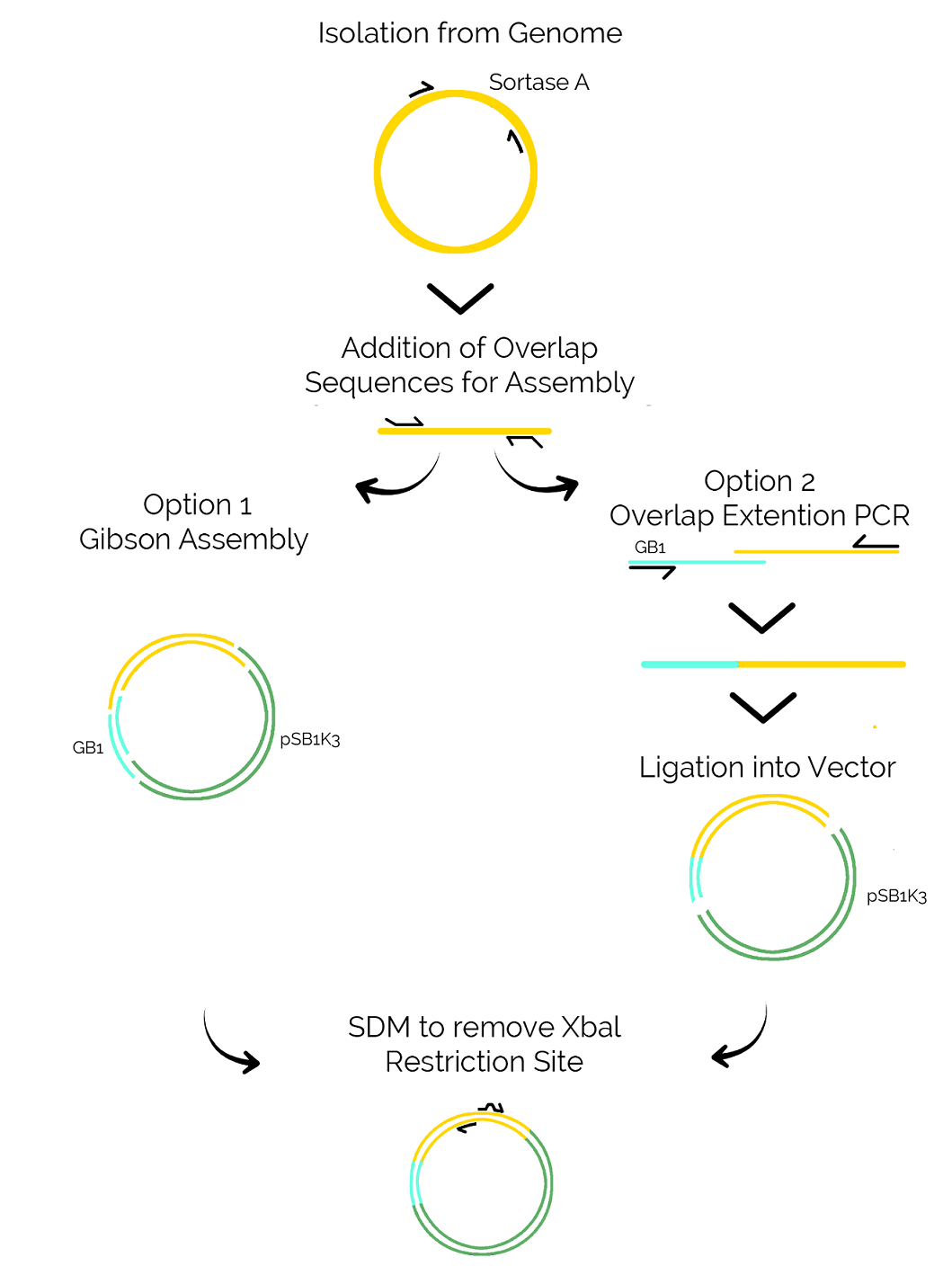
Gibson Assembly relies on the activity of a 5’ exonuclease which creates 3’ overhangs on the complementary sequences, leading to annealing of the two fragments.
Overlap Extension PCR on the other hand requires no exonuclease and instead utilizes a slightly modified PCR reaction. Two different fragments (in our case GB1 and Sortase A) are added into a PCR reaction and will be denatured. In the following step, the DNA strands will be allowed to anneal again and due to the overlapping sequences, a part of the GB1 strands will anneal to Sortase A strand. By carefully selecting the primer combination for this reaction, those recombinant DNA molecules will preferably be amplified and can be used for downstream applications.
Functional Testing Sortase A
After successful assembly with Gibson assembly or overlap extension PCR, we planned to add a T7 promoter to the GB1-SortA hybrid for subsequent expression, purification and functional resting.
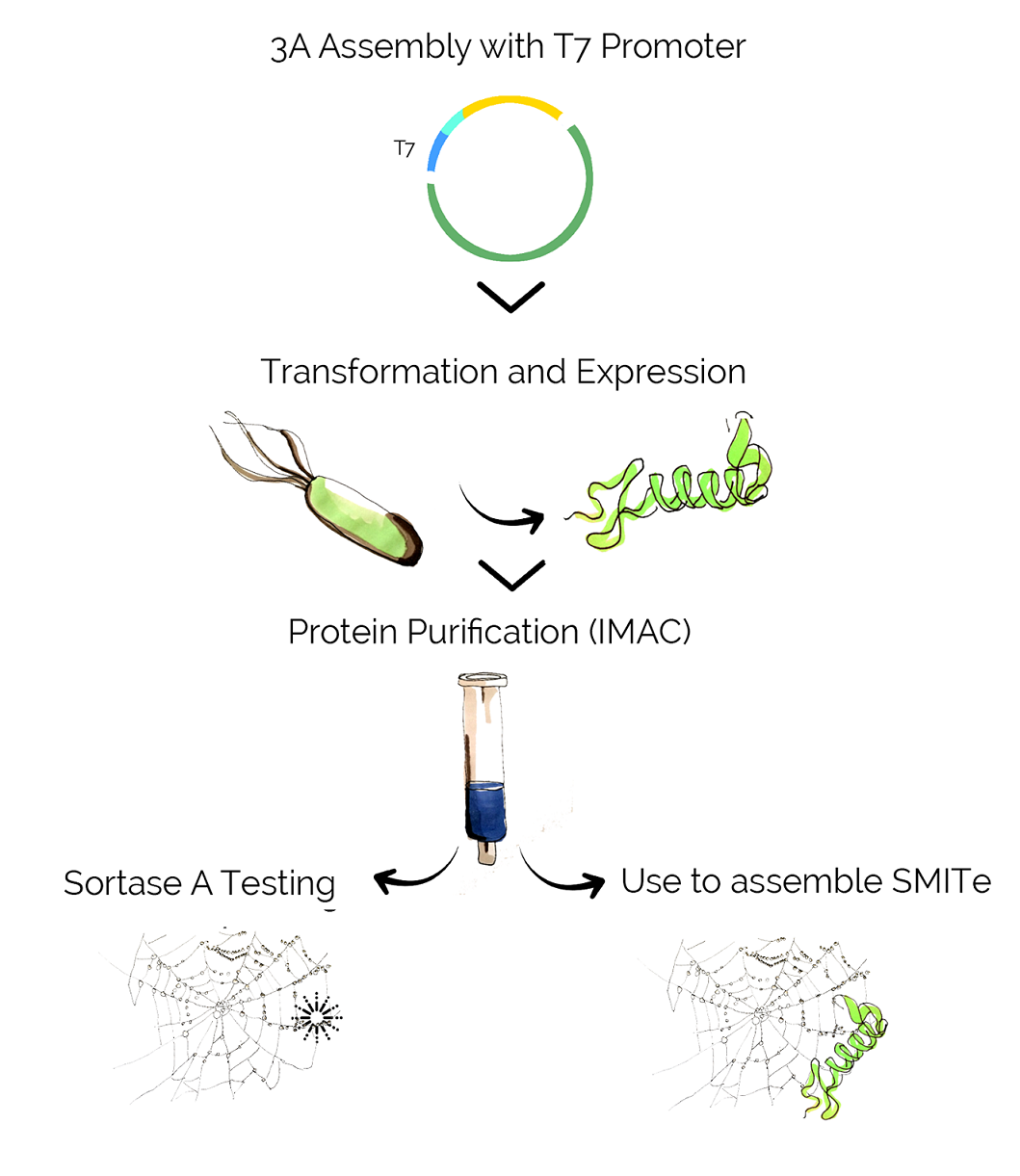
To test the activity of Sortase A, simple methods can be applied: attaching two peptides with the correct tags to each other and performing an SDS-Page to observe the shift in molecular weight. Additionally, fluorescent proteins such as RFP or UnaG can be attached and visualized using a fluorescence microscope to get a better idea of the distribution and density of attached proteins. A final test would of course be the assembly of our finished product, SMITe.
References
‘Firework’ icon by Giuditta Valentina Gentile, arrow icons by Molly Bramlet and Eugen Belyakoff from The Noun Project.


