Defensin
Summary
Defensin (Def) is one of our combat proteins which targets the cell membrane of the bacterial cells. By expressing and attaching it to our spider silk, in combination with the other combat proteins, we hope to combat both biofilm and virulence in chronic wounds.
Assembling a construct for recombinant expression of Def [✔️]
Demonstrating expression and investigating solubility of Def in E. coli [❓]
Demonstrating efficient biofilm inhibition by Def [✔️]
Demonstrating efficient biofilm dispersal by Def [❌]
Demonstrating bacteriolytic activity of Def [❌]
Adding Sortase and His tags to the expression construct [✔️]
Expressing and purifying a construct with added tags [✔️]
Conjugating expressed defensin with added tags to spider silk protein [✔️]
Results
Assembling a construct for recombinant expression of Def
The 3A assembly method is used where the defensin biobrick created by the NYMU Taipei iGEM team 2013 BBa_K1104301 and the T7-promoter created by the Bielefeld Germany iGEM team 2011 BBa_K525998 are cut out from their original constructs and the backbone cut open, using appropriate restriction enzymes.
The ligation products are then first transformed into TOP10 for maximum transformation efficiency. The plasmid are extracted and transformed into BL21(DE3) for protein expression.
3A Assembly, First and Second Attempt [❌]
The first attempt of digesting pSB1C3-Def was unsuccessful and the experiment was simply repeated. In the second attempt, the digestion product was used for ligation, where the gel electrophoresis of the ligation product showed one band at the expected size of 2264 bp. Since the ligation was successful, it was further transformed into both TOP10 and BL21(DE3) cells. Only TOP10 cells colonies were present on the plates. Subsequent extraction and PCR analysis of the plasmid showed clear bands at a size of approximately 300 bp, and no bands at the expected size of approximately 400 bp. Ligation was re-done in order to eliminate any possible random errors.
3A Assembly, Third Attempt [✔️]
The third attempt was successful. However, only the transformation into TOP10 cells were successful. For the expression of defensin, the plasmid had to be re-transformed into BL21(DE3).
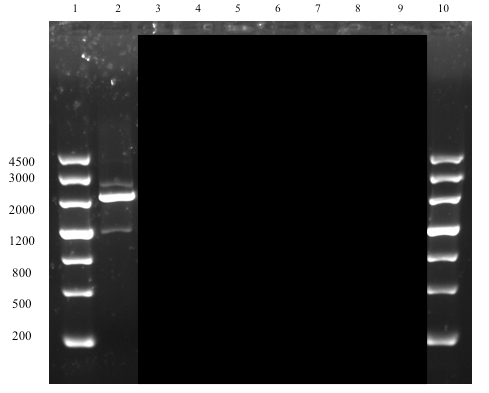
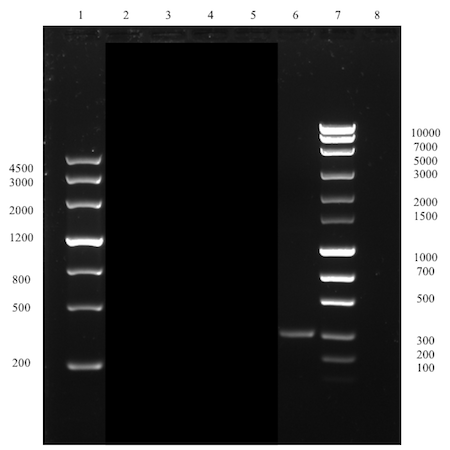
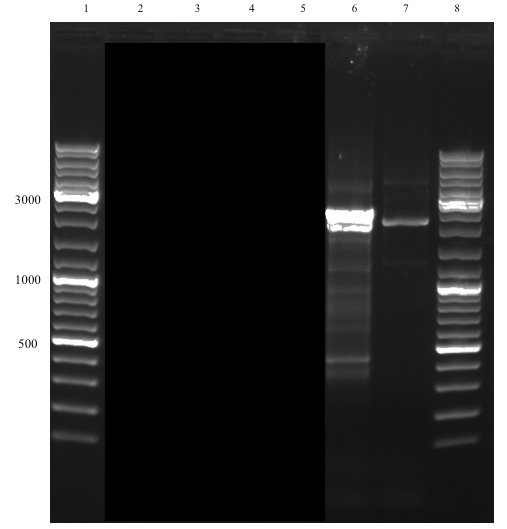
The successful assembly was confirmed with gel electrophoresis of i) the ligation product (see Figure 1, well 14, expected size: 2264 bp), ii) a single digestion of a PCR amplification product of the extracted plasmid after transformation (see Figure 2, well 2, expected size: 2264 bp), and iii) a later performed overhang PCR amplification using the Def_R reverse primer (designed with a BamHI restriction site overhang) instead of the VR (see Figure 3, well 6, expected size: ~300 bp), which all showed clear bands at expected sizes. This part (BBa_K2144003) was submitted to the iGEM Part Registry.
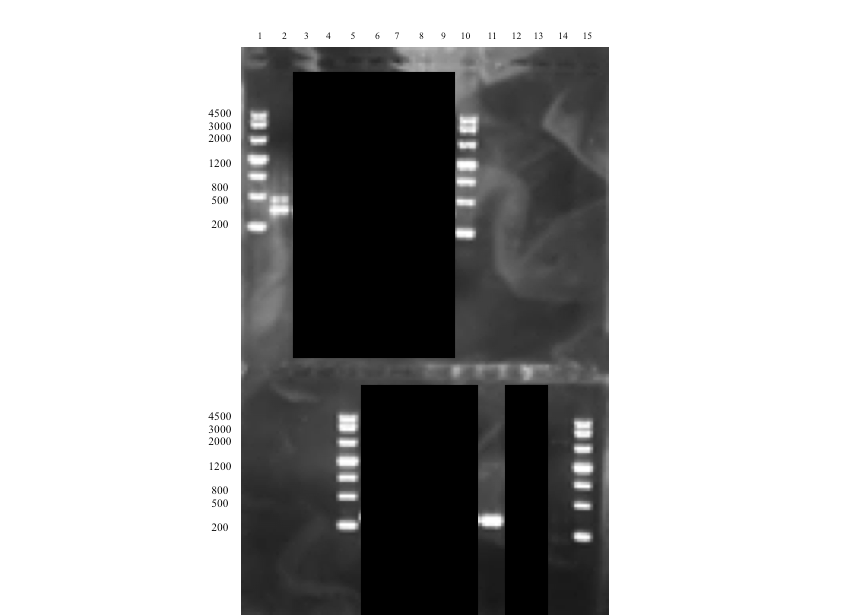
Demonstrating expression and investigating solubility of Def in E. coli
The assembled expression construct are transformed into BL21(DE3) for optimal expression conditions. Liquid cultures of transformed cells are then induced with 0.5 mM or 1 mM IPTG at OD600 0.6 to initiate expression of defensin. After lysing the cells by sonication and collecting the cell debris with centrifugation, both the supernatant and the resuspended pellet are analyzed using SDS-PAGE.
Expression, extraction & SDS-PAGE [❓]
The absence of visible bands corresponding to defensin could be due to a too low concentration of the protein in the cell lysate. However, there could be another reason. Besides bands corresponding to the protein of interest, there is also expected to be a substantial amount of other bands corresponding to the proteins normally present in the bacterial cells. The absence of most of these proteins and defensin indicates that the sonication might have been to harsh and degraded most of the proteins.
Demonstrating efficient biofilm inhibition by Def
A crystal assay was performed to evaluate the inhibition efficiency of defensin on biofilm growth by P. aeruginosa and S. aureus, respectively. Different volumes of cell lysates from BL21(DE3) cells transformed with pSB1C3-T7-Def are tested. Differences between the negative control and Def are then evaluated using Student’s t-test. A p-value <0.05 is considered statistically significant.
Biofilm assay, First Attempt [✔️]
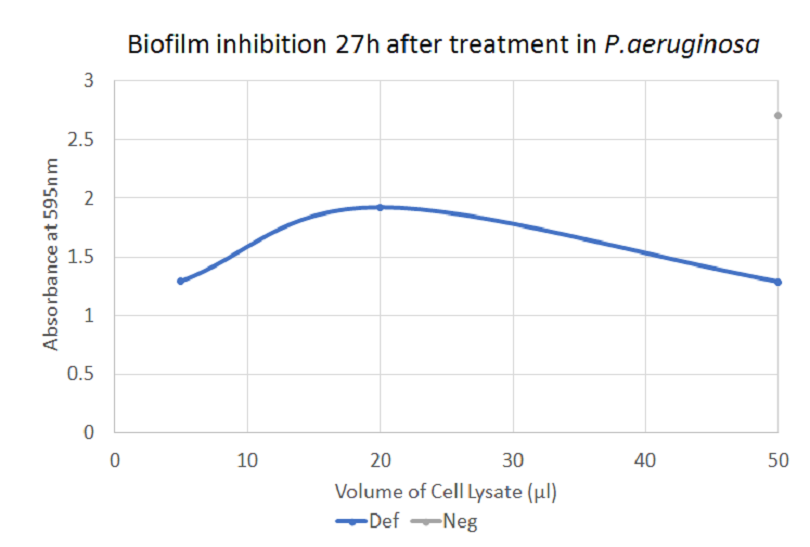
The graph shows how defensin inhibited biofilm formation in P. aeruginosa. There is a significant difference between the negative control and 5 µl and 50 µl of treatment with defensin, respectively.
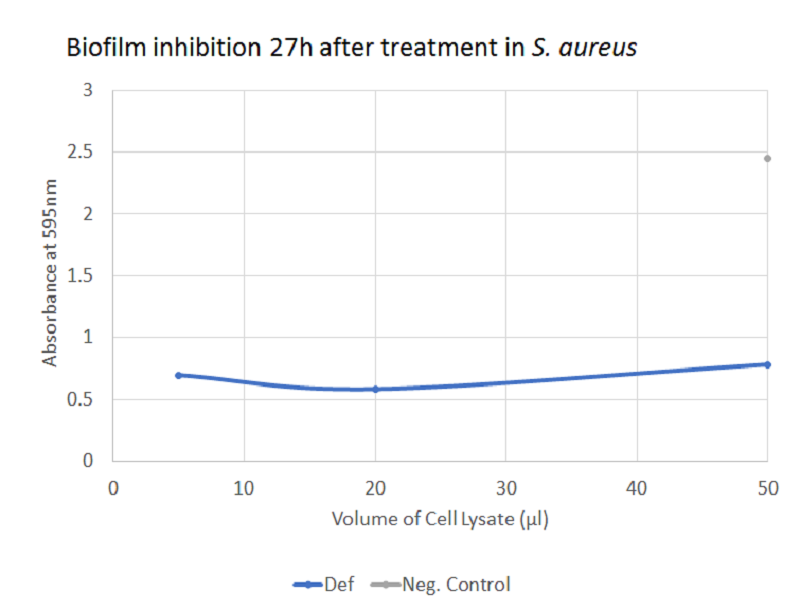
The graph illustrates how defensin inhibited biofilm-formation in S. aureus. There was a significant difference between the negative control and each of the three volumes of treatment with defensin, thus supporting the role of defensin as a potential combat protein for the inhibition of biofilm formation in S. aureus. However, caution has to be taken with the interpretation since crystal violet also stains bacterial cells and not only biofilm - part of the effect observed may be due to a bacteriolytic activity of defension.
Demonstrating efficient biofilm dispersal by Def
To demonstrate biofilm dispersal by defensin, two crystal violet assays are used, one with P. aeruginosa as the biofilm-producer and the other with S. aureus. For the experiment, the already established biofilm is treated with 5 μl, 20 μl or 50 μl of whole cell lysate containing defensin. Differences between the negative control and Def are then evaluated using Student’s t-test. A p-value <0.05 is considered statistically significant.
Biofilm Assay, First Attempt [❓]
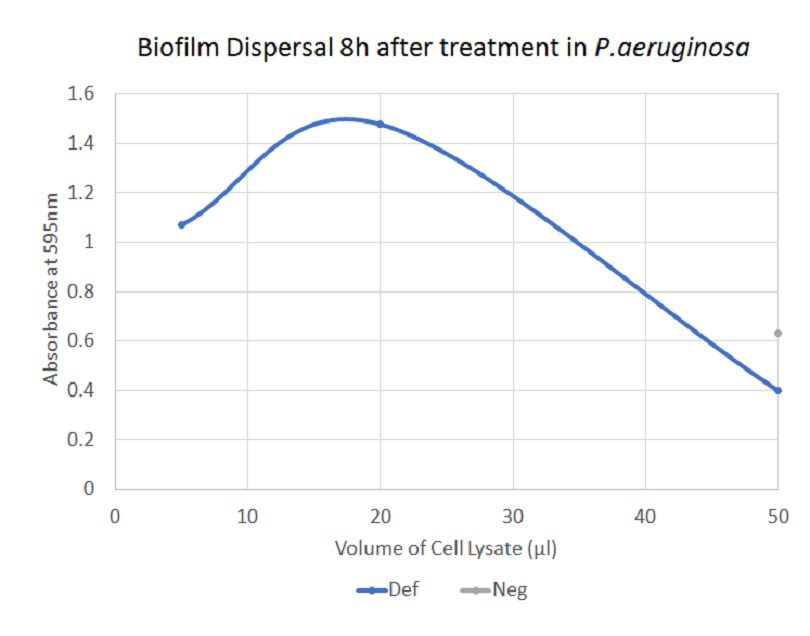
The graph does not show a clear biofilm-dispersing activity by defensin. Whereas the larger volumes of treatment seems to lead to a decreasing absorbance, the same applies to the larger volumes of negative control. This may be due to contamination or due to unspecific effects of the cell debris and thus the results may not be reliable.
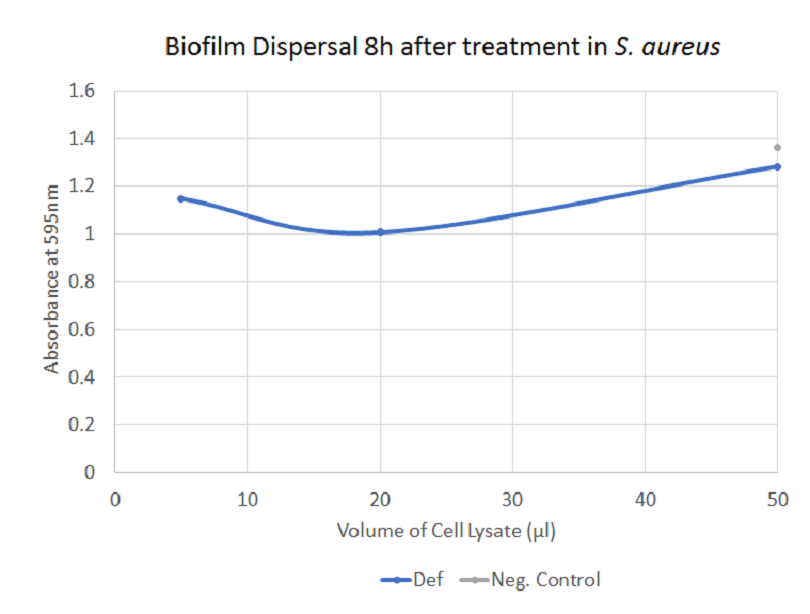
The graph illustrates how defensin (combination of pellet and supernatant) dispersed biofilm by S. aureus. There is a significant difference between negative control and 5 μl and 20 μl of treatment, respectively.
Unfortunately, no clear effect of defensin could be demonstrated - the changes observed after application of the treatment are generally small and do not always correlated with the amount of sample added. Additionally, the negative controls occasionally display an inhibitory effect as well.
Demonstrating bacteriolytic activity of Def
A modified Kirby-Bauer test is used to assess defensin's bacteriolytic activity. TOB1 E.coli are streaked on agar plates onto which filter papers impregnated with the bactericide are placed. Additionally to the cell lysate samples containing the combat protein, a positive controls in the form of an antibiotic is applied as a reference. The bactericidal activity is then measured by observing the “halo” around the impregnated paper.
Kirby-Bauer, First Attempt [❌]
No impeded growth of cells could be seen in the Kirby-Bauer tests; neither from the supernatant samples nor the pellet samples. Most likely, the concentration of defensin in the samples used were too low to show the desired activity on the test. The same samples were used for the biofilm assay, which gave more precise and reliable data regarding the activity of defensin.
Adding Sortase and His tags to the expression construct
When attaching a His-tag and a Sortase A recognition motif (LPETGG) sequence (i.e. linker tag sequence, LT) on the 3’ end of the defensin sequence, the whole pSB1C3-T7-Def plasmid is amplified using Phusion polymerase, DefR as the reverse primer (with BamHI overhang) and SuffixF as the forward primer. The now linearized amplified plasmid with added BamHI restriction site is then double digested with BamHI and PstI and finally ligated in one step to create the pSB1C3-T7-Def-BamHI-LT construct.
Assembly, First attempt [❌]
The overhang PCR amplification of the entire plasmid was unsuccessful. There many unspecific bands at smaller sizes on the gel after electrophoresis, indicated a too low annealing temperature.
Further attempts would be performed at higher annealing temperatures. Also, by adding DMSO, any suspicions regarding difficulties in denaturing the template plasmid, will be eliminated.
Assembly, Second Attempt [✔️]
By increasing the annealing temperature and adding DMSO, the overhang PCR amplification resulted in a clear band at the expected size of 2273 bp with only a few extremely faint unspecifc bands (see Figure 9, well 7). These were neglected and the sample was used for digestion with subsequent ligation with the linker-tag (LT) sequence. The restriction sites of both the linearized plasmid (amplification product) and the LT sequence were located on the very ends of the DNA strands. Thus, no gel electrophoresis was performed to analyse the digestion product since the size of the digested construct would be impossible to compare with the undigested for drawing any conclusions on whether the digestion was successful or not. The digestion product of the two sequences were instead PCR purified and directly ligated.
The successful transformation of the ligation product indicated very strongly that the ligation was successful and that the desired plasmid had been transformed into both TOP10 and BL21(DE3) cells. The assembly of the tag-fused defensin expression construct, was further proven to be successful with successful expression, IMAC purification and conjugation to spider silk protein. This part (BBa_K2144007) was submitted to the iGEM Part Registry.
Expressing and purifying a construct with added tags
As for the pSB1C3-T7-Def construct, a BL21(DE3) E. coli strain is used for the transformation of the pSB1C3-T7-Def-BamHI-LT plasmid for expression. At OD600 0.6, the cells are induced with 0 mM, 0.5, mM, and 1.0 mM IPTG to initiate expression of tag-fused defensin. The cells in the cultures are then lysed and the defensin purified using Immobilized Metal Affinity Chromatography (IMAC). A SDS-PAGE of the eluate is used to determine the presence of the protein of interest.
Purification, First Attempt [❓]
Concentration measurements at 280 nm of the elution fractions indicated presence of protein in the purified sample. However, the SDS-PAGE after IMAC purification did not show any bands at all for the pellet resuspension sample and only very faint bands for the supernatant sample. Probably, the concentrations were too low to be detected - a Western Blot could be used instead but was not performed due to time constraints.
Conjugating expressed defensin with added tags to spider silk protein
The enzyme Sortase A is used to conjugate defensin to spider silk protein. It recognizes the LPETGG recognition motif added on the 3' end of defensin and links it to poly-G sequences on the spider silk protein.
Conjugation, First attempt [✔️]
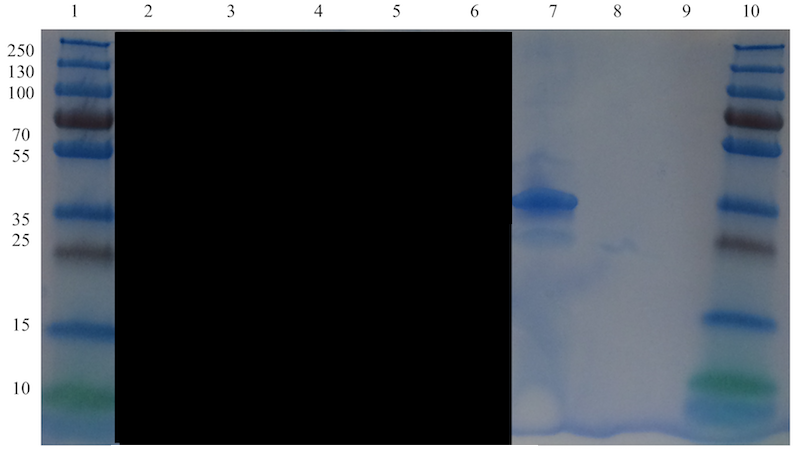
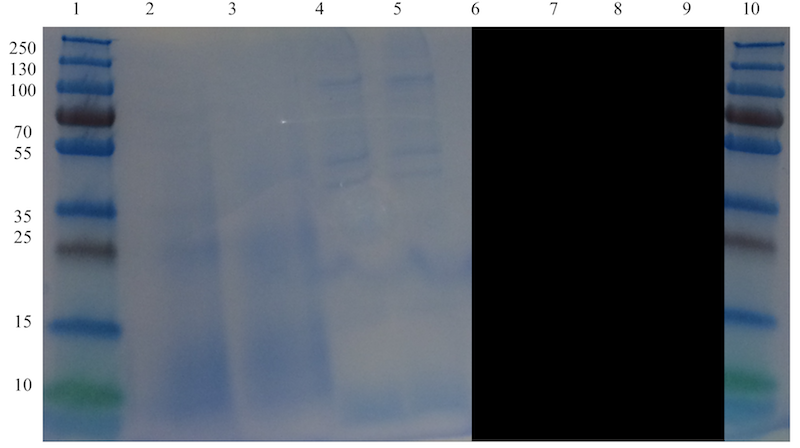
A clear band at the expected size of approximately 33.7 kDa, corresponding to defensin-fused spider silk protein, can be seen in well 7 in Figure 10. A faint band just below that band, at an approximate size of 23-24 kDa is visible and probably corresponds to spider silk protein not fused with defensin. The non-fused spider silk reference in well 9 is not visible, which is most likely due to the very small amount added as the total amount was limited.
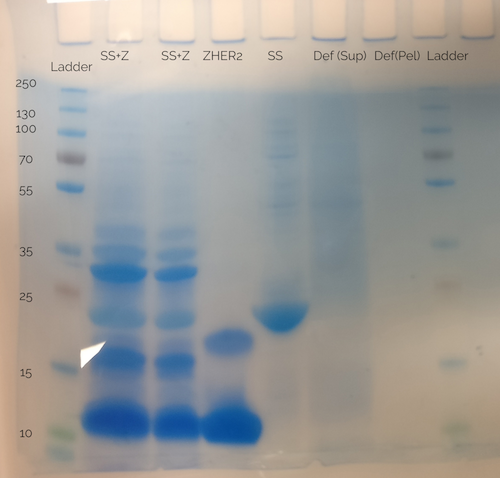
Since the reference in the gel (see Figure 10) is not visible, the spider silk-defensin fusion sample was cross-referenced with another SDS-PAGE made earlier with a non-fused spider silk protein sample (see well labeled "SS" in Figure 11). There, the band corresponding to spider silk protein is clearly at a size of approximately 23 kDa, the expected size of the spider silk protein. From this, the conclusion can be made that the conjugation of recombinant produced defensin, with attached His-tag and LPETGG recognition motif, to spider silk protein using Sortase A, was successful.


