| Line 233: | Line 233: | ||
<ul class="submenu"> | <ul class="submenu"> | ||
<li><a href="https://2016.igem.org/Team:NAU-CHINA/Basic_Part">Basic Part</a></li> | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Basic_Part">Basic Part</a></li> | ||
| − | <li><a href=" | + | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Composite_Part">Composite Part</a></li> |
<li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_Collection">Collection</a></li> | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_Collection">Collection</a></li> | ||
</ul> | </ul> | ||
Revision as of 11:49, 18 October 2016
Bioremediation is considered to be a major method of degrading contaminants in the environment.
We will demonstrate four BioBrick devices in our project. The four devices consist of the device to degrade 3-phenoxybenzoate, 3-hydroxybenzoate,Catechol respectively and a device with the function of thoroughgoing degradation.
Proof of 3-PBA degradation:
3-PBA is degraded as the process shown in figure.1

Figure.1

Figure.2
We structure the device as the figure.2shows.Then it is placed into DH5α.

figure.3 shows that engineered bacteriawhich we used in testing degradation of 3-PBA. We found that some of the solution getting brown for a while. Through figure.4,wewill know that it is the color of the oxideof Catechol solution. Although we have catechol 2,3- dioxygenase, some catecholget into solutionthrough cell membrane making the solution get brown.

Figure.4
In figure.4, A is the bottle withengineered bacteria and 1000μmol/L 3-PBA. Bottle B is the solution of Catechol which has been oxidized. The color of the two bottle is almost the same.

Figure.5
The figure.5 is the engineered bacteria's degradation of 3-PBA tested by HPLC.
From the figure we found that 3-PBA in solution with high concentration is degraded to a certain extent.

Figure.6
The figure shows us 3-PBA's restraint of our engineering bacteria.

Figure.7
The abscissa is the concentration of 3-PBA in each group. The ordinate is the absorbance of bacterial fluid tested by spectrophotometer which can reflect the growth situation of bacteria. The sample we tested comes from the 24th hour's bacterial fluid. In each group from the figure, the left one is the growth situation of our engineering Escherichia coli, the right one comes from data ofSphingobium wenxiniae JZ-1T.
Discussion:
As we can find from Figure.6 , 3-PBA will restrain the growth ofengineering bacteria a lot when its concentration is larger than 1000μmol/L. However, comparing with the growth situation of Sphingobium wenxiniae JZ-1T in same concentration of 3-PBA ,our engineering bacteriacan adapt to high concentration of 3-PBA and Sphingobium wenxiniae JZ-1T couldn't be tolerant ofhigh concentration of 3-PBA.
We think that catechol comes out from the dead cell since cell will let out its inclusion while dying. The darker the solution color is, the more catechol was let out. So that figure.5 and figure.6 can explain the change of color in figure.3. Only the concentration is equal or larger than1000μmol/L can enough catecholbe produced by cells to change the color of solution. Because of the restraint of 3-PBA, the bacteria grew slower as 3-PBA's concentration rises. Therefore our engineered bacteria in solution with 1000μmol/L 3-PBA can produce a lot of Catechol and let it out much more than other groups since more bacteria were dead than in solution with 1500μmol/L and 2500μmol/L of 3-PBA. As a result, the middle bottle in figure 3 has the darkest color.
Degradation of 3-Hydroxybenzoate
3-Hydroxybenzoate (3-HBA) is degraded as the processshown in figure.7 . Besides, we need MhbT proteinto let E.coli absorb 3-HBA from extracellular domain.
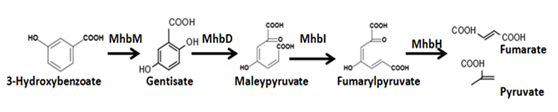
Figure.8

Figure.9 device structure

Figure.10
We placed the device into DH5ɑ and testedits function, the result shows in the figure.9.The ordinateis the peak area tested by high efficiency liquid chromatography(HPLC). From the figure we will find that the 3-HBA can be degraded thoroughlywithin 24 hours. And the concentration below 5000μmol/L can be degraded thoroughly within 12 hours.
Proof ofCatechol degradation

Figure.11
Catechol is degraded as the way shown in figure.10.

Figure.12
We structure the device as the figure.11 shows.Then it is placed into DH5ɑ.

Figure.13
Figure.12 shows a plate with each three streaks of three different colonies, "J23100 + B30034 + C23O + dT" and "J23100 + C23O(with native RBS) + dT"are colonies transformed with C23O coding gene, while negative control(CK) is avector without C23O gene. Before dripping catechol solution onto the colonies, the three streaks showed the same color colonies just like what CK shows on the plate.After dripping 0.2mol/L catechol solution onto the colonies, the color of "J23100 + B30034 + C23O + dT" and "J23100 + C23O(native RBS) + dT" colonies turned to bright yellow, which suggest that our C23O geneis able to work ,for 2-hydroxymuconate semialdehyde in figure.10 shows bright yellow. It can also demonstrate the device shown in figure.11 works.[2]
Proof of thoroughgoingdegradation

Figure.14
We structure the device as the figure.13 shows.Then it is placed into DH5ɑ. The big device contains all of the three devices above.
The whole system was placed on two different plasmids J61002and PCC1.

Figure.15
Copy number control to tune gene expression:
We use pCC1 vector which is a CopyControl vectors. On LB chloramphenicol plates or in LB media supplemented with chloramphenicol, CopyControl clones grown in TransforMax EPI300 E. coli replicate at single-copy number from the F-factor replicon because expression of the trfA gene is repressed. Addition of CopyControl Induction Solution induces the cells to express the trfA gene product and induces their replication at high-copy number from oriV. Replication from oriV results in higher yields and higher purity of cloned DNA. The strains we use is TransforMax EP1300 Chemically competent which lack the tonA gene and are engineered for use with CopyControl Cloning Systems. The cells contain an inducible mutant trfA gene whose gene product is required for initiation of replication from the oriV origin of replication.
As the figure.2 shown, section A is on the PCC1 and section B is on J61002, for J61002 and PCC1 have different antibiotic resistance . We used the bacteria with PCC1 making competent cell, then transfer plasmid J61002 with section B into a PCC1 competent cell, using dual resistance to pick out the bacteria with two plasmids.
When testing the degradation, we added two kinds of antibiotics(Ampicillin and Chloramphenicol)into the medium to prevent the loss of plasmids. we added copper ion into medium(50μmol/Lfinal concentration of copper ion), to induce copA promoter.

Figure.16
As figure.3 shows, our engineered bacteria can degrade about 15-20% of 3-PBA within 24 hours. For two kind of antibiotics have been added into the medium, our engineered bacteria will grow much slower than common bacteria. It may explain our engineered bacteria's slow speed of degrading 3-PBA.

Figure.17
The abscissa is the concentration of 3-PBA in each group. The ordinate is the absorbance of bacterial fluid tested by spectrophotometer which can reflect the growth situation of bacteria. The sample we tested comes from the 12th hour's bacterial fluid. In each group from the figure, the left one is the growth situation of JZ-1, the middle one comes from the data of CK, the right one comes from data of our engineered bacteria.
The figure shows us the different growth situation betweenSphingobium wenxiniaeJZ-1T, ourengineering bacteria and CK(E.coli DH5α). From the figure, we can find that our engineering bacteria can tolerate higher concentration of 3-PBA than Sphingobium wenxiniaeJZ-1T. Also, E.coli can tolerate higher concentration of 3-PBA than Sphingobium wenxiniaeJZ-1T.In the future, we will connect the whole device to a single vector and realize more efficient degradation.

Figure.18
The figure shows the degradation of our engineered bacteria which contains the whole device above.
It shows that our engineered bacteria can work in high concentration, and about 20% of 3-PBA in solution can be degraded in 24 hours.
The whole device was demonstrated to work, and the concept of degrading is also demonstrated.
References
[1]Xu Y, Gao X, Wang S H, et al. MhbT is a specific transporter for 3-hydroxybenzoate uptake by Gram-negative bacteria[J]. Applied and environmental microbiology, 2012, 78(17): 6113-6120.
[2] He Z. Novel organization of catechol meta pathway genes in the nitrobenzene degrader Comamonas sp. JS765 and its evolutionary implication. Journal of industrial microbiology & biotechnology, 2007, 34(2): 99-104.


