Part Collection
The Sortase Package, is a generalized solution, designed to enable conjugation/immobilization of any protein of interest to a fusion partner with an exposed glycine residue. The Sortase A package consists of three key parts:
Sortase A BioBrick: A Biobrick encoding for Sortase-A.
Linker-tag (LT) sequence: A custom designed linker-tag sequence encoding for the Sortase A recognition Motif, LPETGG, and a Histidine tag for purification purposes (fig.1).
Protein primer design solution: A Primer designed to anneal downstream the protein coding sequence with a BamHI overhang.
Sortase A BioBrick - BBa_K2144008
This Biobrick encodes for a truncated version of the transpeptidase Sortase A, fused with the Protein G B1 domain (GB1) acting as a solubility tag.
Linker-Tag - BBa_K2144101
The linker-tag sequence has a BamHI restriction site upstream its sequence. Digesting the BamHI and standard suffix PstI site, enables a compatible ligation of the LT sequence with any protein of interest when used in combination with the “protein primer design solution” and the vector BackBone suffix PstI site.
 Figure 1. An illustration of the main features in the custom designed linker-tag sequence.
Figure 1. An illustration of the main features in the custom designed linker-tag sequence.
Protein Primer Design Solution
Designing primers for use with our Vector containing the Linker-Tag Sequence (LT) for Sortase A might be easier with a few pointers:
- The forward strand of your protein should be coding so that the C-terminus of your protein is fused to the linker-tags. Sortase A conjugates C-terminal LPXTG to N-terminal poly-G sequences.
- Make sure to avoid including the stop codons or terminators typically used at the C-terminal of coding constructs as they will interrupt translation of the LT downstream of your protein.
- The primers have to be carefully tested since some of them require the insertion of an extra base before the BamHI restriction site in order to avoid a frameshift between protein and LT.
- Include a few extra bases on the 5’ end – BamHI does not digest efficiently at the very end of DNA molecules.
A primer we designed for our lysostaphin BioBrick can be observed Below.
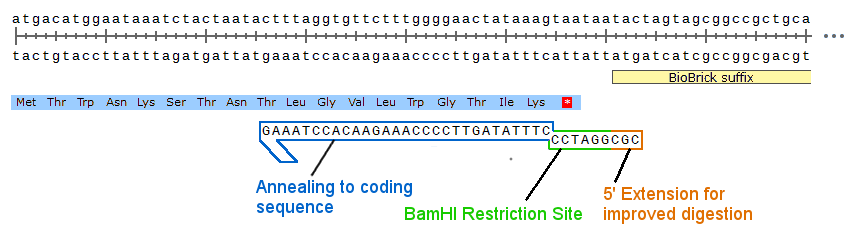
By generalizing our Sortase package, we have designed a system in which future iGEM teams can easily utilize Sortase A to conjugate/immobilize any type of fusion partners of interest.


