Proof of Concept
Introduction
The main goal of our project is to demonstrate biofilm dispersing activity of combat proteins that has been conjugated to spider silk by the help of sortase. The final approach of our proof of concept remains to be tested, however we have managed to show aspects of our project goal. There are three steps to our goal.
-Step 1: Prove that the combat proteins posses antagonistic activity towards tested strains and possibly inhibit and disperse biofilms.
-Step 2: Prove that Sortase work as a conjugation enzyme between two fusion components, where one of the component contains an expressed sortase recognition motif that is fused to the protein sequence.
-Step 3: Prove that the product, combat proteins conjugated to spider silk by using Sortase, is capable of disrupting biofilm.
So how did we test this?
We have used two different analysis methods for step 1. The two methods are called Kirby-Bauer test and Crystal Violet Biofilm assay. A more detailed procedure of how these methods work are described below. For step 2 we have used SDS-PAGE to analyze the conjugation reaction of sortase. If smaller proteins would have been used in the conjugation, then HPLC or MS would have been an option to analyze the subsequent result. Unfortunately, we have no current results for Step 3. We can however perceive the result of Step 3 as a combination of Step 1 and Step 2. Since we have the results for the first two steps, we can to certain extent implicate the results we have to project expected results of Step 3.
Kirby-Bauer test
This method is a fast procedure to determine the effect of the cell lysate on the growth of tested culture on an agar plate. E.coli TOB1 cells which is a biofilm forming strain was used in this test. The procedure begins by first spreading the liquid culture on to McConkey agar. Subsequently, filter papers soaked in cell lysate containing one of the combat proteins are placed on the agar and incubated with the culture. As the TOB1 cells starts to grow, the cell lysate will either inhibit or allow the cells to grow around the paper depending on the biofilm-degrading capability of the combat protein in the cell lysate. If the combat protein can inhibit or degrade cell growth there will halos will be observed around the paper where no bacteria has grown. As a positive control, paper soaked in kanamycin was used. This is the same principle as testing if a certain bacterial strain has developed antibiotic resistance or not.
Biofilm assay (96-well plate)
The second method used to proof our goal of demonstrating biofilm-degrading proteins is a Crystal Violet Biofilm assay. In this assay, cultures of S.aureus or P.aeruginosa are grown in a 96-well microtitre plate and later subjected to treatment using cell lysates that contain combat proteins. When the biofilm has been formed, the excess cells are removed and the biofilm is stained with crystal violet 0.1% staining solution. The degree of staining is quantified using a photometric analysis at 595 nm. A change or loss in color of the well from dark purple indicates a successful degradation of the biofilm. Crystal violet works by binding to cellular components, thus an effective treatment would result in the reduced amount of crystal violet bound to the biofilm. In our inhibition test, cell lysates containing our combat protein were added to the culture-containing wells and analysed after 48 hours. A similar procedure to determine biofilm dispersal was also performed by adding the cell lysates to established biofilms instead. As a positive control, gentamicin was used which is known to degrade biofilm.
Lysostaphin
Our combat protein lysostaphin have shown positive results with both methods and was especially effective towards S. aureus. Figure 1 shows the result from the Kirby Bauer test where halos are visible. The induced cells shows a larger halo than the non-induced cells, indicating that lysostaphin is capable of lytic activity. The same result was apprehended in the biofilm assay where one of the two lysostaphin samples gave a clear decrease in absorbance while the negative control remained the same. Lysostaphin also effectively dispersed the pre-existing biofilm, as shown in the Figure 3. More of these results can be seen in the lysostaphin results page, section “Demonstrating efficient Biofilm dispersal by lysostaphin” and “Demonstrating efficient Biofilm inhibition by lysostaphin”.
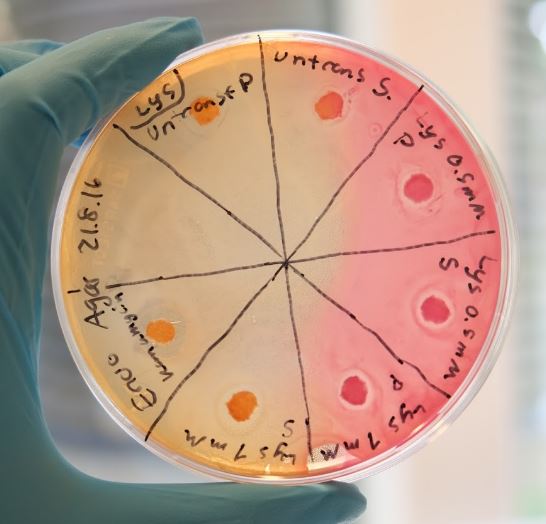

 Figure 3. Biofilm dispersal test of Lysostaphin on P.aeruginosa 8 hours after treatment
Figure 3. Biofilm dispersal test of Lysostaphin on P.aeruginosa 8 hours after treatment
Esp
Esp detaches biofilm by cleaving cell wall anchored proteins of S. aureus. Our synthesised EB showed bactericidal activity and potent biofilm dispersal capacity. In the Kirby Bauer tests of Esp, clear halos can be seen in right sample of Figure 4. The left figure display halos indicating bactericidal activity, but in these test, the proteins used were frozen thus this could have led to the smaller sized halos. Esp showed biofilm dispersing qualities in the Biofilm assay, which was as efficient as the positive control in Figure 5. Further results can be seen on the results page under section “Investigating possible bactericidal activity of Esp”

 Figure 5. Biofilm dispersion test of Esp on S. aureus
Figure 5. Biofilm dispersion test of Esp on S. aureus
Nuc
The antibacterial results for Nuc were found to be negative. This was expected as Nuc targeted exposed nucleic acids which was a component of the biofilm, rather than that of free living cells. In the biofilm dispersal assay, Nuc showed some activity. Figure 6 confirms this finding by showing a decreased absorbance with increased sample volume.

Def
Defensin has showed positive results in both the Kirby-Bauer tests and the Biofilm assay. The result from the Biofilm assay is displayed in Figure 7. The figure showed a significant decline in the absorbance of crystal violet was observed compared to the negative control. Further experiments and results can be seen in the results page, section “Demonstrating efficient Biofilm dispersal by Defensin”.

Sortase conjugation
In the section “Lab -> Lab book -> Sortase week 16” the conjugation between spider silk and a small peptide called Protein ZHER was conjugated by using Sortase. The Sortase in this experiment was kindly given to us by one of our supervisors, Kristina Westerlund. This experiment was done to prove that the enzyme Sortase works as a catalyzer of a conjugation reaction between two fusion partners with a LPETGG-tag and a exposed glycine residue, respectively. The figure below shows the SDS-PAGE with samples from the conjugation. Well 2 and 3 is the product and well 4 and 5 is the negative control. The bands visible between 30-45 kDa in well 2 and 3 is the conjugated product.

Summary and the future of our project
Based on the results, we have at least two combat proteins which have shown efficient biofilm degrading qualities in two different tests. We have shown that a conjugation between a protein and spider silk using sortase is also fast and efficient.
Demonstrating that our Sortase Biobrick can conjugate combat proteins to spider silk and degrade biofilms still remains as future works. Nonetheless, we have seen speed progression and several positive results with defensin. Had it that we were not limited on time, we believe that defensin has the potential of being conjugated to spider silk and possibly degrade biofilms.


