| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{NCKU_Tainan | + | <html><head> |
| − | < | + | <meta charset="utf-8"><link rel="shortcut icon" href="/wiki/images/8/80/T--NCKU_Tainan--favicon.png" type="image/x-icon"><link rel="icon" type="image/png" href="/wiki/images/8/80/T--NCKU_Tainan--favicon.png"><link rel="icon" type="image/x-icon" href="/wiki/images/8/80/T--NCKU_Tainan--favicon.png"><meta name="viewport" content="width=device-width, initial-scale=1.0"><meta property="og:title" content="Project Description - iGEM NCKU"><meta property="og:site_name" content="Project Description - iGEM NCKU"><meta property="og:description" content=""><title>Project Description - iGEM NCKU</title><meta http-equiv="Content-Type" content="text/html" charset="utf-8"><meta property="og:image" content=""><meta property="og:image:type" content="image/png"><link rel="stylesheet" href="/Team:NCKU_Tainan/css/frame/T--NCKU_Tainan--bootstrap_min_css?ctype=text/css&action=raw"><link href="/Team:NCKU_Tainan/font/T--NCKU_Tainan--NotoSans_css?ctype=text/css&action=raw" rel="stylesheet" type="text/css"><link rel="stylesheet" href="/Team:NCKU_Tainan/font/T--NCKU_Tainan--font-awesome_min_css?ctype=text/css&action=raw"> <link rel="stylesheet" href="/Team:NCKU_Tainan/css/T--NCKU_Tainan--Layout_css?ctype=text/css&action=raw"> |
| + | </head> | ||
| + | <body> | ||
| + | <style>@font-face { font-family: 'NotoSansCJKtc-Regular'; src: url("/wiki/images/0/0b/T--NCKU_Tainan--NotoSansCJKtc-Regular.woff") format('woff');}</style><nav class="navbar navbar-default"><div id="navbar-container" class="container-fluid"> <div class="navbar-header"> <button type="button" class="navbar-toggle collapsed" data-toggle="collapse" data-target="#navbar" aria-expanded="false"> <span class="sr-only">Toggle navigation</span> <span class="icon-bar"></span> <span class="icon-bar"></span> <span class="icon-bar"></span> </button> <a class="navbar-brand" href="/Team:NCKU_Tainan"> <h1>NCKU</h1><h4>Tainan</h4> </a> </div> <div id="navbar" class="navbar-collapse collapse"> <ul class="nav navbar-nav"> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu1" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="">Project</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu2"> <li><a href="/Team:NCKU_Tainan/Project">Background</a></li> <li><a href="/Team:NCKU_Tainan/Description">Description</a></li> <li><a href="/Team:NCKU_Tainan/Results">Results</a></li> <li><a href="/Team:NCKU_Tainan/Model">Modeling</a></li> <li><a href="/Team:NCKU_Tainan/Parts">Parts</a></li> </ul> </li> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu2" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="">Device</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu2"> <li><a href="/Team:NCKU_Tainan/Hardware">Hardware</a></li> <li><a href="/Team:NCKU_Tainan/Software">Software</a></li> <li><a href="/Team:NCKU_Tainan/Demonstrate">Demonstrate</a></li> </ul> </li> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu3" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="">Judging</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu3"> <li><a href="/Team:NCKU_Tainan/Medal">Medal</a></li> <li><a href="/Team:NCKU_Tainan/Safety">Safety</a></li> </ul> </li> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu4" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="/Team:NCKU_Tainan/Team">Team</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu4"> <li><a href="/Team:NCKU_Tainan/Team">Team</a></li> <li><a href="/Team:NCKU_Tainan/Attributions">Attributions</a></li> <li><a href="/Team:NCKU_Tainan/Acknowledgement">Acknowledgement</a></li> <li><a href="/Team:NCKU_Tainan/Collaborations">Collaborations</a></li> </ul> </li> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu5" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="/Team:NCKU_Tainan/Human_Practices">Human Practices</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu5"> <li><a href="/Team:NCKU_Tainan/Human_Practices">Overview</a></li> <li><a href="/Team:NCKU_Tainan/Integrated_Practices">Integrated Practices</a></li> <li><a href="/Team:NCKU_Tainan/Engagement">Engagement</a></li> </ul> </li> <li> <a class="dropdown-toggle" type="button" id="dropdownMenu6" data-toggle="dropdown" aria-haspopup="true" aria-expa="" nded="true" href="">Notebook</a><div class="nav-underline"></div> <ul class="dropdown-menu" aria-labelledby="dropdownMenu6"> <li><a href="/Team:NCKU_Tainan/Notebook_Construction">Construction</a></li> <li><a href="/Team:NCKU_Tainan/Notebook_Functional_Test">Functional Test</a></li> <li><a href="/Team:NCKU_Tainan/Notebook_Device_Design">Device Design</a></li> <li><a href="/Team:NCKU_Tainan/Notebook_Protocols">Protocols</a></li> <li><a href="/Team:NCKU_Tainan/Notebook_Model">Model</a></li> </ul> </li> </ul> <ul class="nav navbar-nav navbar-right"> </ul> </div><!--/.nav-collapse --></div><!--/.container-fluid --></nav><div id="container-big"><div id="iGEMbar"></div><div id="line-left"></div><div id="line-left2"></div><div id="line-right"></div><div id="photo-left"></div></div><!--/.container-big --> <style> | ||
| + | #photo-left { background-image: url("/wiki/images/f/f3/T--NCKU_Tainan--Description.png"); } | ||
| + | </style> | ||
| + | <div class="container-fluid" style="margin-top:100px"> | ||
| + | <div class="head">PROJECT / Description</div> | ||
| + | <div class="content row"> | ||
| + | <div class="col-md-9"> | ||
| + | <div class="head2">U-KNOW - Our Project</div> | ||
| + | <div class="title-line" id="overview">Overview</div> | ||
| + | <p>Diabetes is a chronic and lifelong disease found in many people. This disease can cause serious consequences when people are not aware of it. without careful attention and proper care, it could develop one of its potentially life-threatening complications. To be treated properly, regular glucose monitoring can serve as a basis for diagnosis and management of diabetes. Also, it is clinically proved that a healthy person with attention to the blood sugar control, lifestyle modifications and medications can successfully avoid the complications associated with it., Monitoring the sugar would be a good start for glucose control and achieve the aim, prevention. There are many diabetic self-monitoring tools, such as test strips and meters. The majority of their users are diabetic users adopting them for good sugar control, but not really widely used for prevention. Moreover, most of the blood glucose measuring devices on the market are invasive types and not user friendly. That is why we raised the idea for our device, U-KNOW.</p> | ||
| + | <p>Our device, U-KNOW, is a noninvasive tool using a colorimetric method that decodes the color response shown on a urine glucose measuring strip. This method can be routinely used in health examinations as an warning indicator for diabetes. This portable monitoring device contains a biosensor to measure urine glucose noninvasively. By reading the urine sample, the glucose level can be shown on the device and on the smart devices, including smart phone and wearables. The U-KNOW system avoids the unpleasant pain and also improves the limitations of the type using the colorimetric method.</p> | ||
| + | <p>This synthetic biological circuit device was created by <a href="/Team:NCKU_Tainan/Hardware">low cost eletronic parts and is a light-weight instrument</a> designed for self-monitoring of urine glucose level. The users are only required to put a few drops of urine in to our disposable column to process the measurement. The result, then, can be transferred to your phone or smart devices from a home health care monitoring system through a bluetooth. Additionally, the home health care monitoring system collect glucose data to generate statistical reports and graphs that produce a comprehensive graph showing the changes of the user’s recent glucose level.</p> | ||
| − | <div class=" | + | <div class="title-line" id="detect">Detection of Glucose</div> |
| − | < | + | <p>In this part, we plan to detect the glucose concentration via bacteria. We designed this biosensor for urine glucose measurements via a plasmid integrating a glucose-induced promoter PI (<a href="http://www.idf.org/membership/wp/taiwan">BBa_K861170</a>) and a monomeric RFP (<a href="http://parts.igem.org/Part:BBa_E1010">BBa_E1010</a>). And there are significantly positive correlation between fluorescence intensity and glucose concentrations.</p> |
| − | < | + | <p>Promoter PI consists of overlapping consensus cAMP-receptor-protein-binding sites (CRP-binding sites) and consensus RNA polymerase binding site[1]. Hence, the steric hindrance between CRP and RNA polymerase will repress downstream genes at a high concentration of CRP[2]. In <i>E. coli</i> strains (e.g.DH5α and Bl21), cAMP and glucose present a negative correlation. The increasing adenylyl cyclase activities increases the amount of cAMP. It causes a decrease of glucose concentration and a higher CRP concentration.[3]. cAMP will bind the CRP which, in its bound form, can specifically bind the consensus CRP-binding site of Pl promoter, leading to repression of downstream genes. In contrast, when the glucose concentration is high, the adenylyl cyclase activity, cAMP, and CRP are decreased in the bound form. Consequently, an more explicit expression of downstream RFP E1010 can be a reporter gene as well as an indicator of the presence of glucose.</p> |
| − | <p> | + | <img src="/wiki/images/5/5c/T--NCKU_Tainan--projDes1.jpg"> |
| − | + | ||
| − | + | ||
| − | </ | + | |
| − | <div class=" | + | <div class="title-line" id="safety">Safety System</div> |
| − | < | + | <p>In terms of bio-safety, we designed the circuit that consists of P<sub>BAD</sub> promoter and lysis gene. P<sub>BAD</sub> is an <i>E. coli</i> promoter induced by L-arabinose. In the absence of arabinose, the dimerized repressor protein araCs would bind to the AraI1 operator site of P<sub>BAD</sub> and the upstream operator site AraO2 would block transcription of P<sub>BAD</sub>. With the presence of arabinose, AraC (<a href="http://parts.igem.org/Part:BBa_I13458">BBa_I13458</a>) dimers would bind to it and then change its conformation so that it interacts with the AraI1 and AraI2 operator sites, permitting the downstream transcription[4]. Using P<sub>BAD</sub> as the promoter for lysis empowers us to control the lysis process better. However, under the environment of a high glucose concentration, P<sub>BAD</sub> would be definitely inhibited. Thus, the P<sub>BAD</sub> expression system can be controlled with two often-seen substance, glucose and arabinose. Yet, in order to make glucose be effective to P<sub>BAD</sub> the required concentration should exceed 0.01%.</p> |
| − | < | + | <div class="img"> |
| − | < | + | <img src="/wiki/images/1/1b/T--NCKU_Tainan--projDes2.png"> |
| − | </div> | + | <div>Arabinose is added under the test paper coated with our <i>E. coli</i></div> |
| + | </div> | ||
| + | <p>Moreover, in our device, powder of L-arabinose is added at the bottom of column, which would not permeate the membrane until the urine is added. After adding the urine sample, the L-arabinose would then dissolve and react with <i>E. coli</i>, which is coated on the test paper. If the sample contains no glucose at all, the circuit will be turn on to produce lysis which result in cytolysis (or cell death)[5].</p> | ||
| + | <div class="title-line" id="ref">Reference</div> | ||
| + | <p>[1] B De Crombrugghe, et al., Cyclic AMP receptor protein: role in transcription activation, Science (1984)</p> | ||
| + | <p>[2] S Busby, et al., Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein, Journal of molecular biology (1982)</p> | ||
| + | <p>[3] M Lavigne, et al., Transcription activation by cAMP receptor protein (CRP) at the Escherichia coli gal P1 promoter. Crucial role for the spacing between the CRP binding site and the -10 region, Biochemistry (1992)</p> | ||
| + | <p>[4] NL Lee, et al., Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and P<sub>BAD</sub>, in the L-arabinose regulatory region of Escherichia coli, PNAS (1984)</p> | ||
| + | <p>[5] L.M. Guzman, et al., Tight regulation, modulation, and high-level expression by vectors containing the arabinose P<sub>BAD</sub> promoter, Journal of Bacteriology (1995)</p> | ||
| + | </div> | ||
| + | <div class="col-md-3"> | ||
| + | <ul id="sidemenu" style="width:90%;"> | ||
| + | <li><a href="#overeview" onclick="return toEvent('overview');">Overview</a></li> | ||
| + | <li><a href="#detect" onclick="return toEvent('detect');">Detection of Glucose</a></li> | ||
| + | <li><a href="#safety" onclick="return toEvent('safety');">Safety System</a></li> | ||
| + | <li><a href="#ref" onclick="return toEvent('ref');">Reference</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> <!-- /.container-fluid --> | ||
| + | <script>document.getElementById("line-left2").style.display="none";</script> | ||
| + | <script src="/Team:NCKU_Tainan/js/frame/T--NCKU_Tainan--jquery-1_12_0_min_js?ctype=text/javascript&action=raw"></script><script src="/Team:NCKU_Tainan/js/frame/T--NCKU_Tainan--bootstrap_min_js?ctype=text/javascript&action=raw"></script><script src="/common/MathJax-2.5-latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML"></script><script src="/Team:NCKU_Tainan/js/T--NCKU_Tainan--MathjaxConfigIgem_js?ctype=text/javascript&action=raw"></script><script src="/Team:NCKU_Tainan/js/T--NCKU_Tainan--common_js?ctype=text/javascript&action=raw"></script><script>(function() { /* change icon */ var link = document.createElement('link'); link.type = 'image/x-icon'; link.rel = 'shortcut icon'; link.href = '/wiki/images/8/80/T--NCKU_Tainan--favicon.png'; document.getElementsByTagName('head')[0].appendChild(link);}());</script> | ||
| − | </html> | + | |
| + | </body></html> | ||
Latest revision as of 16:08, 19 October 2016
Diabetes is a chronic and lifelong disease found in many people. This disease can cause serious consequences when people are not aware of it. without careful attention and proper care, it could develop one of its potentially life-threatening complications. To be treated properly, regular glucose monitoring can serve as a basis for diagnosis and management of diabetes. Also, it is clinically proved that a healthy person with attention to the blood sugar control, lifestyle modifications and medications can successfully avoid the complications associated with it., Monitoring the sugar would be a good start for glucose control and achieve the aim, prevention. There are many diabetic self-monitoring tools, such as test strips and meters. The majority of their users are diabetic users adopting them for good sugar control, but not really widely used for prevention. Moreover, most of the blood glucose measuring devices on the market are invasive types and not user friendly. That is why we raised the idea for our device, U-KNOW.
Our device, U-KNOW, is a noninvasive tool using a colorimetric method that decodes the color response shown on a urine glucose measuring strip. This method can be routinely used in health examinations as an warning indicator for diabetes. This portable monitoring device contains a biosensor to measure urine glucose noninvasively. By reading the urine sample, the glucose level can be shown on the device and on the smart devices, including smart phone and wearables. The U-KNOW system avoids the unpleasant pain and also improves the limitations of the type using the colorimetric method.
This synthetic biological circuit device was created by low cost eletronic parts and is a light-weight instrument designed for self-monitoring of urine glucose level. The users are only required to put a few drops of urine in to our disposable column to process the measurement. The result, then, can be transferred to your phone or smart devices from a home health care monitoring system through a bluetooth. Additionally, the home health care monitoring system collect glucose data to generate statistical reports and graphs that produce a comprehensive graph showing the changes of the user’s recent glucose level.
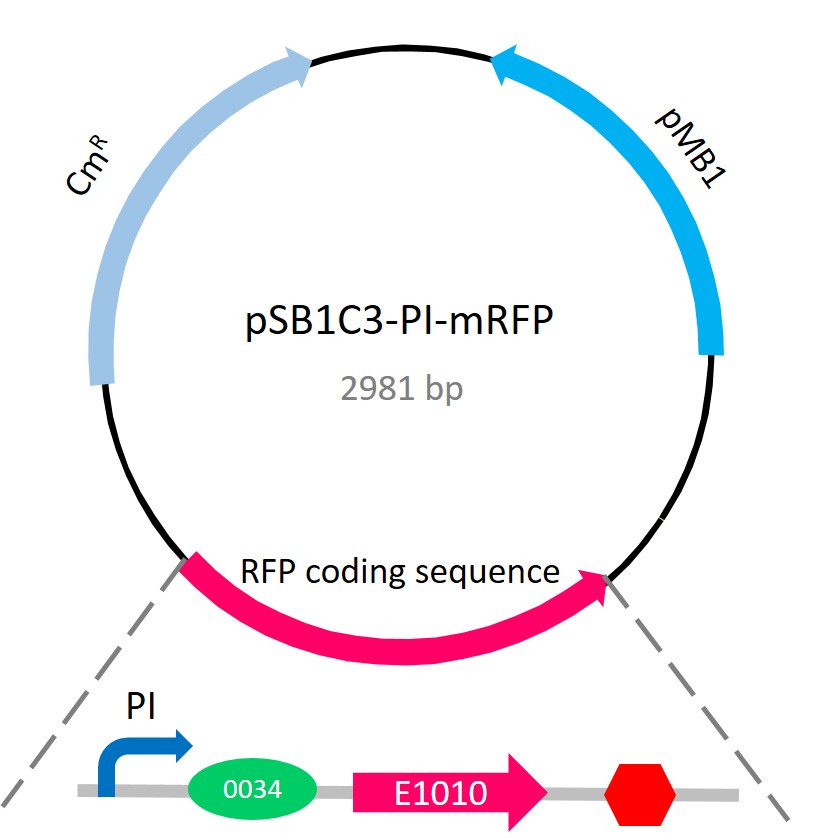
In this part, we plan to detect the glucose concentration via bacteria. We designed this biosensor for urine glucose measurements via a plasmid integrating a glucose-induced promoter PI (BBa_K861170) and a monomeric RFP (BBa_E1010). And there are significantly positive correlation between fluorescence intensity and glucose concentrations.
Promoter PI consists of overlapping consensus cAMP-receptor-protein-binding sites (CRP-binding sites) and consensus RNA polymerase binding site[1]. Hence, the steric hindrance between CRP and RNA polymerase will repress downstream genes at a high concentration of CRP[2]. In E. coli strains (e.g.DH5α and Bl21), cAMP and glucose present a negative correlation. The increasing adenylyl cyclase activities increases the amount of cAMP. It causes a decrease of glucose concentration and a higher CRP concentration.[3]. cAMP will bind the CRP which, in its bound form, can specifically bind the consensus CRP-binding site of Pl promoter, leading to repression of downstream genes. In contrast, when the glucose concentration is high, the adenylyl cyclase activity, cAMP, and CRP are decreased in the bound form. Consequently, an more explicit expression of downstream RFP E1010 can be a reporter gene as well as an indicator of the presence of glucose.
In terms of bio-safety, we designed the circuit that consists of PBAD promoter and lysis gene. PBAD is an E. coli promoter induced by L-arabinose. In the absence of arabinose, the dimerized repressor protein araCs would bind to the AraI1 operator site of PBAD and the upstream operator site AraO2 would block transcription of PBAD. With the presence of arabinose, AraC (BBa_I13458) dimers would bind to it and then change its conformation so that it interacts with the AraI1 and AraI2 operator sites, permitting the downstream transcription[4]. Using PBAD as the promoter for lysis empowers us to control the lysis process better. However, under the environment of a high glucose concentration, PBAD would be definitely inhibited. Thus, the PBAD expression system can be controlled with two often-seen substance, glucose and arabinose. Yet, in order to make glucose be effective to PBAD the required concentration should exceed 0.01%.
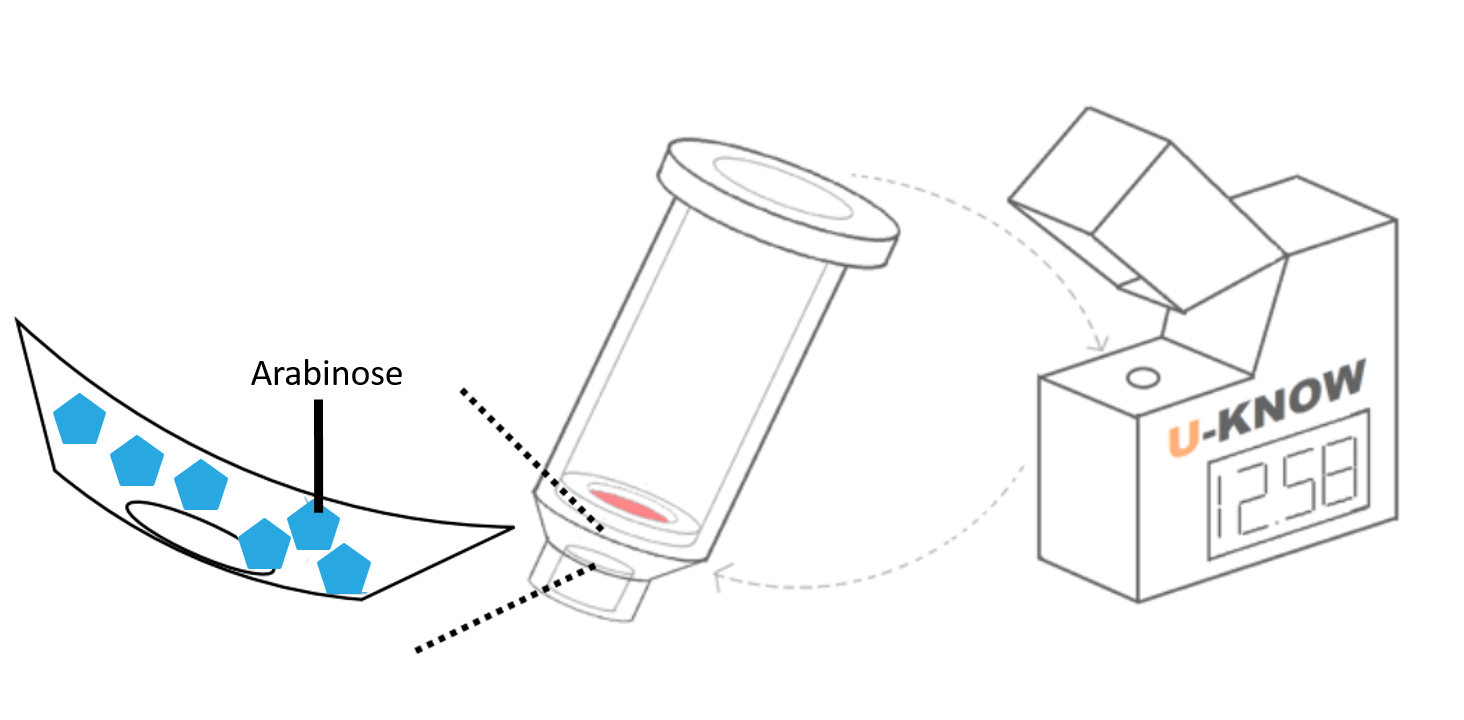
Moreover, in our device, powder of L-arabinose is added at the bottom of column, which would not permeate the membrane until the urine is added. After adding the urine sample, the L-arabinose would then dissolve and react with E. coli, which is coated on the test paper. If the sample contains no glucose at all, the circuit will be turn on to produce lysis which result in cytolysis (or cell death)[5].
[1] B De Crombrugghe, et al., Cyclic AMP receptor protein: role in transcription activation, Science (1984)
[2] S Busby, et al., Mutations in the Escherichia coli operon that define two promoters and the binding site of the cyclic AMP receptor protein, Journal of molecular biology (1982)
[3] M Lavigne, et al., Transcription activation by cAMP receptor protein (CRP) at the Escherichia coli gal P1 promoter. Crucial role for the spacing between the CRP binding site and the -10 region, Biochemistry (1992)
[4] NL Lee, et al., Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli, PNAS (1984)
[5] L.M. Guzman, et al., Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter, Journal of Bacteriology (1995)

