| Line 241: | Line 241: | ||
<li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_List">Part List</a></li> | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_List">Part List</a></li> | ||
<li><a href="https://2016.igem.org/Team:NAU-CHINA/Basic_Part">Basic Part</a></li> | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Basic_Part">Basic Part</a></li> | ||
| − | <li><a href=" | + | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Composite_Part">Composite Part</a></li> |
<li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_Collection">Collection</a></li> | <li><a href="https://2016.igem.org/Team:NAU-CHINA/Part_Collection">Collection</a></li> | ||
</ul> | </ul> | ||
Revision as of 00:36, 19 October 2016
What is 3-hydroxybenzoate(3-HBA)?
3-hydroxybenzoate is the secondary metabolite of 3-phenoxybenzoate which can be degraded sequentially.Mhb proteins are involved in 3-hydroxybenzoate catabolism.We insert the gene cluster into our engineering bacteria to degrade 3-HBA, thus leading the degradation thoroughly without accumulation of metabolite. To test the function of gene cluster MhbDHIM independently, MhbT which acts as achannel proteinis needed to let 3-HBA into cells.

Fig.1.The structure of 3-HBA
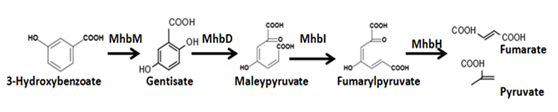
Fig.2. The degradation pathway of 3-hydroxybenzoatet

Fig.3. The growth ofKlebsiella pneumoniae M5a1 in different concentration of 3-hydroxybenzoate
We tested the tolerance ofKlebsiella pneumoniae M5a1 to 3-hydroxybenzoate. From figure.3 we foundthat 8500 and 7500μmol/L 3-hydroxybenzoatecan inhibit the growth of Klebsiella pneumoniae M5a1 completely.
HPLC(High Performance Liquid Chromatography) test
We set a serious of solutions with different concentration of 3-hydroxybenzoate to test the degradation of M5a1 and our engineering bacteria.

Fig.4. Abscissais the concentration of 3-HBA in original medium. Ordinate is the peak area of 3-HBA in medium tested by HPLC.Each group has two columns, the left one shows M5a1`s degradation and the right one shows engineering bacteria`s degradation.
Result
From the figure.4 we can find that our engineering bacteriawill degrade 3-HBA much faster than M5a1 in 24h. Although the engineering bacteria's degradation for high concentration is slower than M5a1 in 12h, it showed efficient ability of degradation within 24h. We supposed that the high concentration of 3-HBA can affect the expression of enzyme inour engineering bacteria. So it cannot degrade 3-HBA well at the beginning. Once the concentrationof 3-HBA reduces in cells, 3-HBA will be degraded quickly.


