In medicine, when the presence of urine glucose exceeds 5mM/L, it implies pre-diabetes or diabetes. However, we refer to people whose urine glucose exceeds 120mM/L as sever diabetic patients. Consequently, finding the person whose urine glucose concentration is over 5mM or 120mM is our target for prevention and early detection.
First, we prove that there is a difference between 0.1mM and 5mM (120mM) in the paired-difference T test part. And, we use the regression and prediction intervals to distinguish exceeding 120mM and 5mM from exceeding 0.1mM. From the result, 5mM can be distinguished from 0.1mM after 101 minutes, and 120mM can be distinguished from 0.1mM/L after 88 minutes.
In medicine, the urine glucose exceeds 5mM implies having diabetes, and it exceeding 120mM implies being a severe patient. Hence, we use paired-difference test to analyze whether the difference of these two groups (0.1 vs 5mM or 0.1 vs 120mM) have statistical significance in this part.
Procedure
1. Predicting if there is a difference over 90 minutes, we analyze the data of 0.1mM and 5mM at 90 minutes first.
(Data)
Experiment Types |
First | Second | ... | Twelfth |
|---|---|---|---|---|
| 0.1mM | 358 | 368 | ... | 338 |
| 5mM | 369 | 386 | ... | 385 |
Because the experiments of 0.1mM and 5mM have correlation and they are small sample, we choose the paired-difference test to examine whether there is difference in two groups at 90 minutes.
Analysis:
(use one-tailed test)
\(H_0:\mu_0\le0\ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ H_\alpha:\mu_0\ge0\)
Because the sample is small, we choose the t distribution to test.
| d(=5mM-0.1mM) | 11 | 18 | ... | 47 |
\(n\) (Number of paired differences)=12
\(\bar{d}\) (Mean of the sample differences)= 26.16666667
\(s_d\) (Standard deviation of the sample differences) = \(\sqrt{\frac{\sum(d_i-\bar{d})^2}{n-1}}\) = 13.19664788
Test statistic t = \(\frac{\bar{d}-0}{s_d/\sqrt{n}}\) = 6.868713411 \(\gt t_{0.05,11}\) = 1.795885
P(t\(\gt\)6.868713411)= 1.995084e-05
Hence, we conclude that there is a difference between 0.1 and 5mM.
2. Find the minimal time at which there is statistical significance by paired-difference test.(use R program & α=0.05)
(Data)
Types Time(min) |
0.1mM | ... | 0.1mM | 5mM | ... | 5mM |
|---|---|---|---|---|---|---|
| 0 | 295 | ... | 276 | 292 | ... | 291 |
| 1 | 296 | ... | 276 | 290 | ... | 291 |
| 2 | 301 | ... | 277 | 292 | ... | 287 |
| . . . |
. . . |
... | . . . |
. . . |
... | . . . |
| 119 | 392 | ... | 372 | 420 | ... | 443 |
| 120 | 392 | ... | 374 | 427 | ... | 439 |
By using R program
\(t_{0.05,11}\) = 1.795885
statistic Time(min) |
T statistic (\(t_0\)) |
|---|---|
| 0 | 2.283227 |
| 1 | 1.992688 |
| . . . |
. . . |
| 119 | 11.908155 |
| 120 | 15.538544 |
Hence, we can find \(t_0 \gt t_{0.05,11}\) at every time.
data0.1<-read.csv("C:/Users/Rick/Desktop/NCKUactivity/iGEM/819test/0.1urine.csv",header=T)
data5<-read.csv("C:/Users/Rick/Desktop/ NCKUactivity/iGEM/819test/5urine.csv",header=T)
difference<-cbind(data5[,2]-data0.1[,2],data5[,3]-data0.1[,3],data5[,4]-data0.1[,4],data5[,5]-data0.1[,5],data5[,6]-data0.1[,6],data5[,7]-data0.1[,7],data5[,8]-data0.1[,8],data5[,9]-data0.1[,9],data5[,10]-data0.1[,10],data5[,11]-data0.1[,11],data5[,12]-data0.1[,12],data5[,13]-data0.1[,13])
dbar<-difference%*% rep(1/12,12)
var<- ((difference -rep(dbar, 12))^2%*%rep(1,12))/11
sd<-var^0.5
t<-(dbar/sd)*(12^0.5)3. Same with procedure 2, find the minimal time at which there is statistical significance by paired-difference test. (use R program, α=0.05, remove a outlier data)
By using R program
\(t_{0.05,10}\) = 1.812461
statistic Time(min) |
T statistic (\(t_0\)) |
|---|---|
| 0 | 4.720629 |
| 1 | 4.580426 |
| . . . |
. . . |
| 119 | 12.428324 |
| 120 | 14.240245 |
Hence, we can find \(t_0 \gt t_{0.05,10}\) at every time.
Conclusion:
We can say there are the differet groups (0.1 vs 5 mM or 0.1 vs 120mM) in part 1 with the data. And then, we need to find an accurate value which can distinguish between two groups.
data0.1<-read.csv("C:/Users/Rick/Desktop/ NCKUactivity/iGEM/819test/0.1urine.csv",header=T)
data120<-read.csv("C:/Users/Rick/Desktop/ NCKUactivity/iGEM/819test/120urine.csv",header=T)
difference<-cbind(data120[,2]-data0.1[,2],data120[,3]-data0.1[,3],data120[,4]-data0.1[,4],data120[,5]-data0.1[,5],data120[,6]-data0.1[,6],data120[,8]-data0.1[,8],data120[,9]-data0.1[,9],data120[,10]-data0.1[,10],data120[,11]-data0.1[,11],data120[,12]-data0.1[,12],data120[,13]-data0.1[,13])
dbar<-difference%*% rep(1/11,11)
var<- ((difference -rep(dbar, 11))^2%*%rep(1,11))/10
sd<-var^0.5
t<-(dbar/sd)*(11^0.5)Our product uses the fluorescence intensity to obtain the concentrations of urine glucose. According to our results, we precisely distinguished the concentrations of 5mM and 120mM from 0.1mM by using fluorescence intensity with 99% accuracy. Consequently, we use the skill of the prediction interval in this part.
Procedure
1. Use the regression to find the model of 0.1, 5, 120mM, and show the prediction interval plot.
(0.1mM)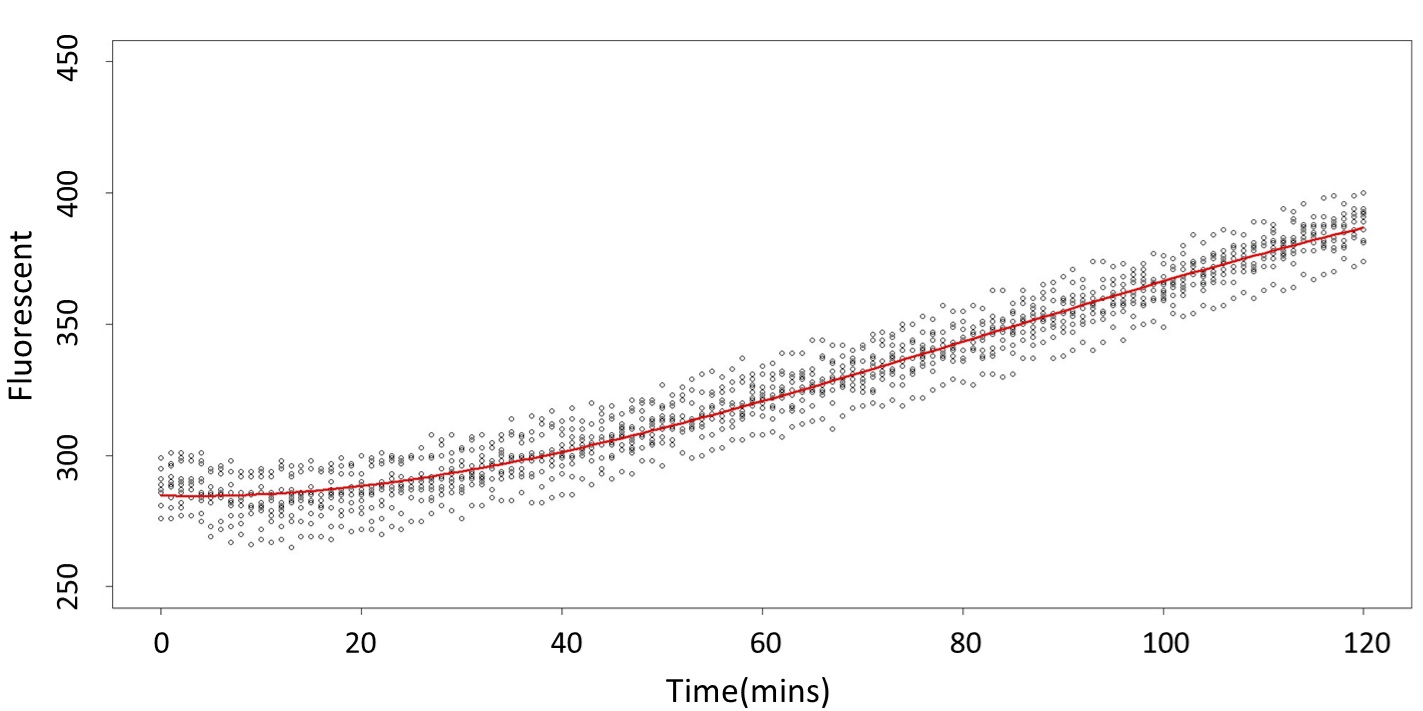
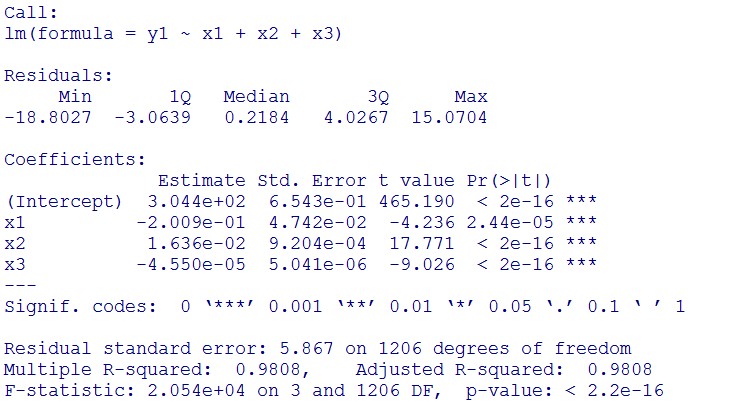
(summary)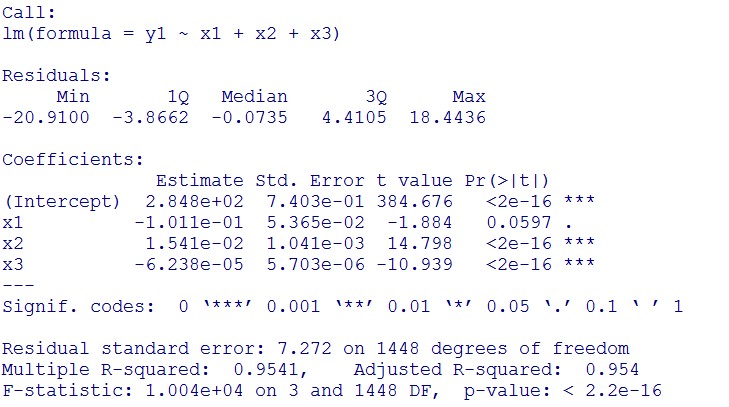
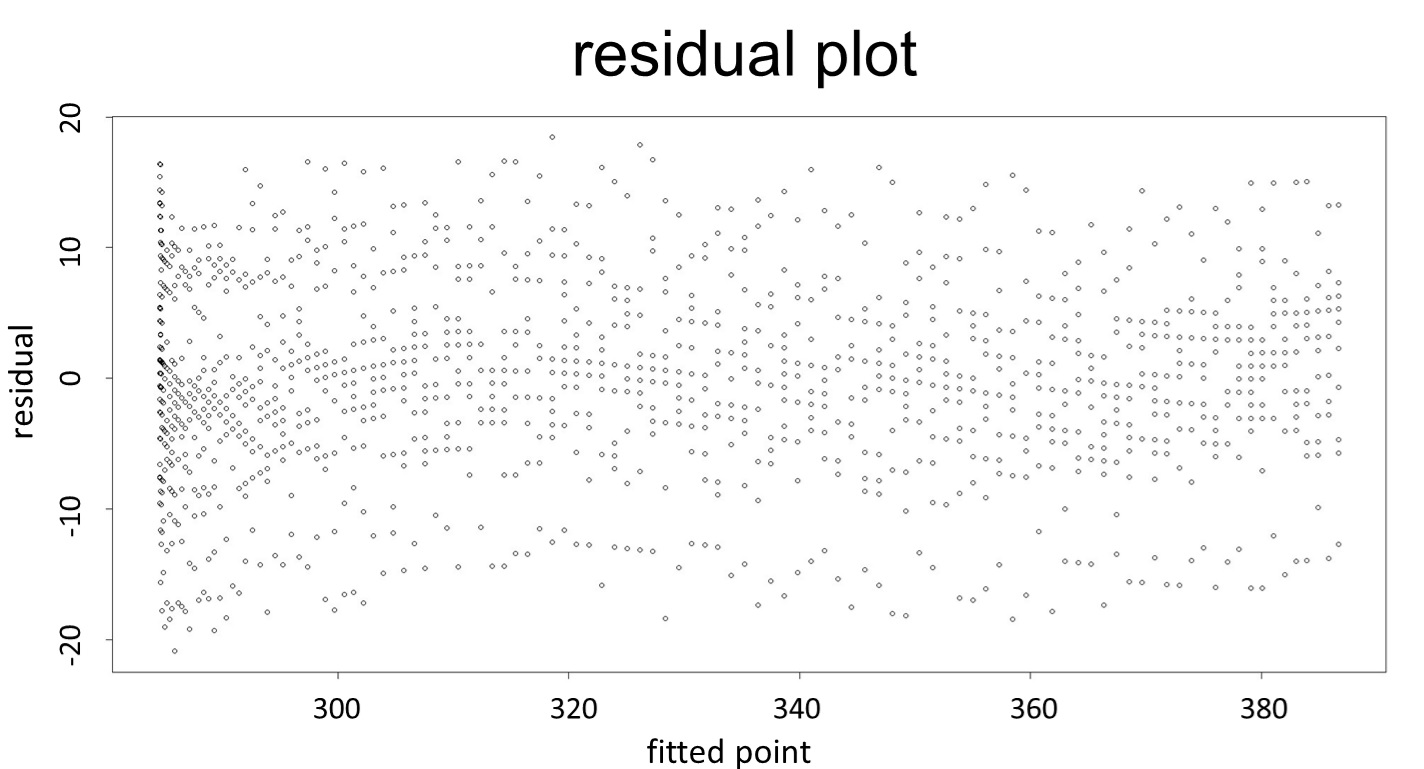
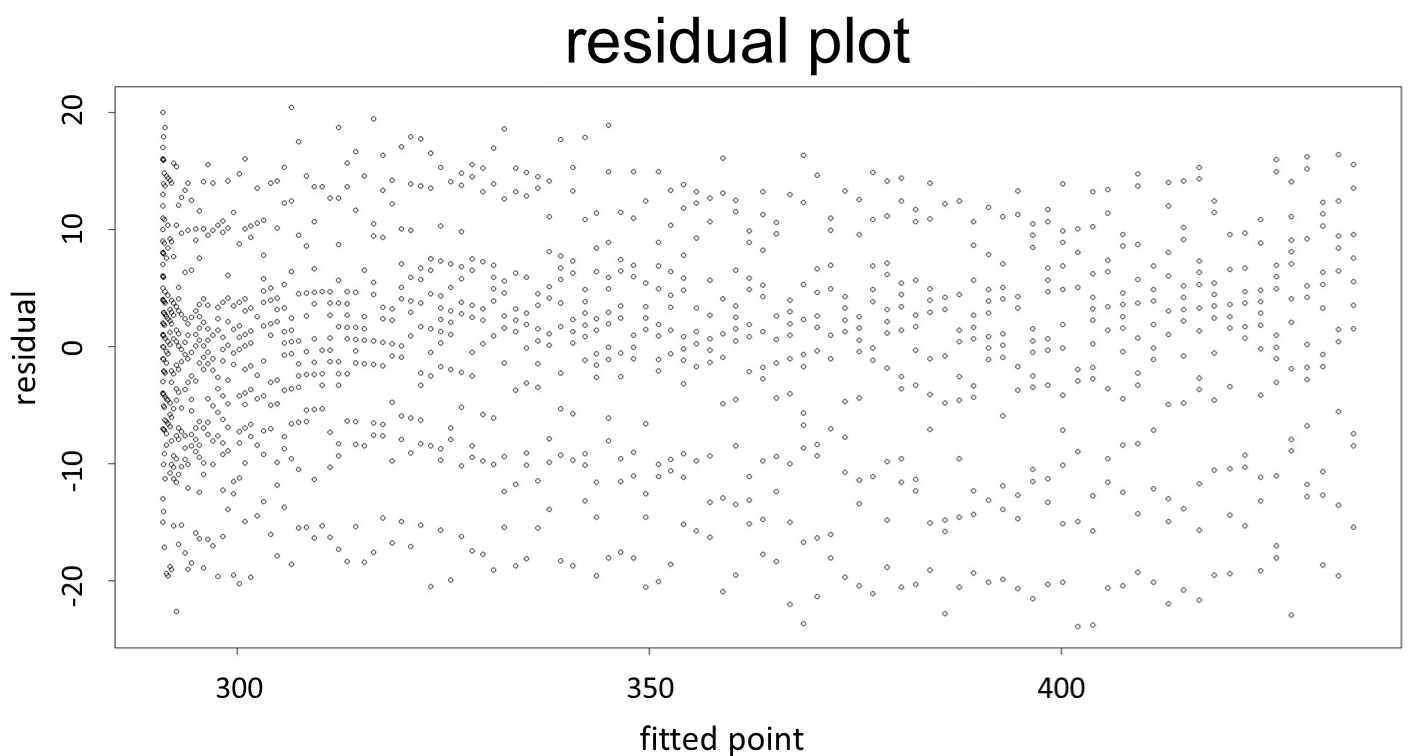
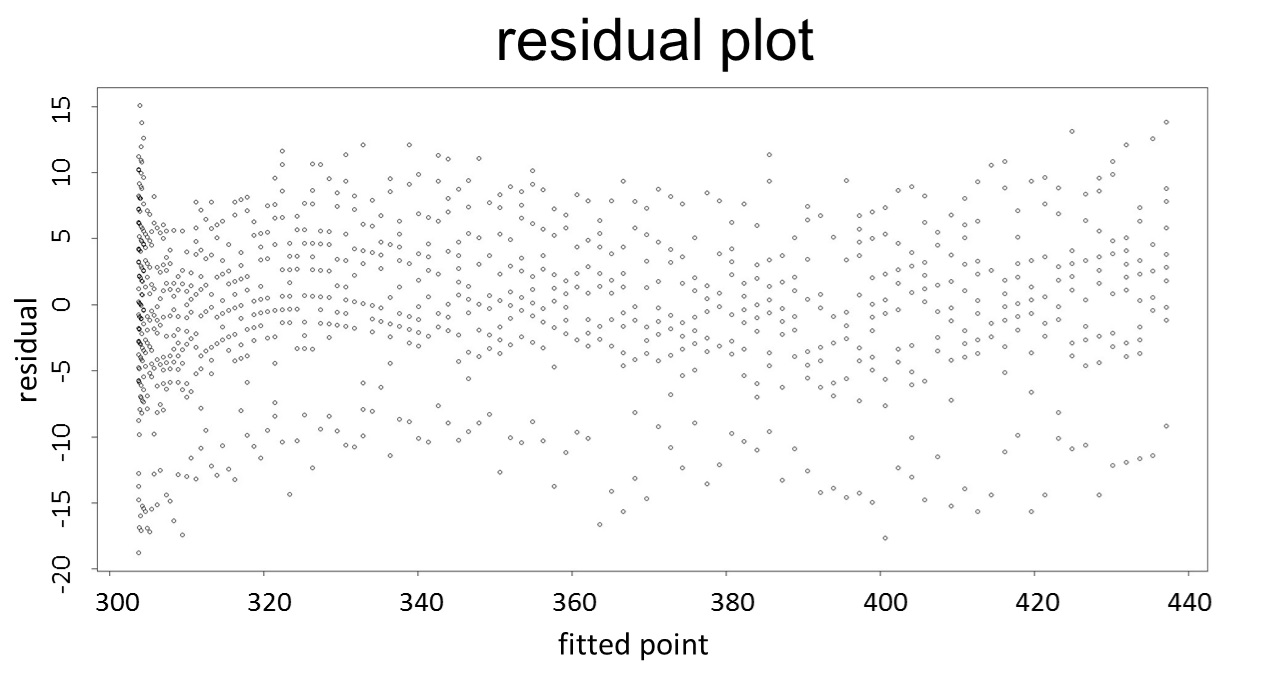
(residual plot) [2]
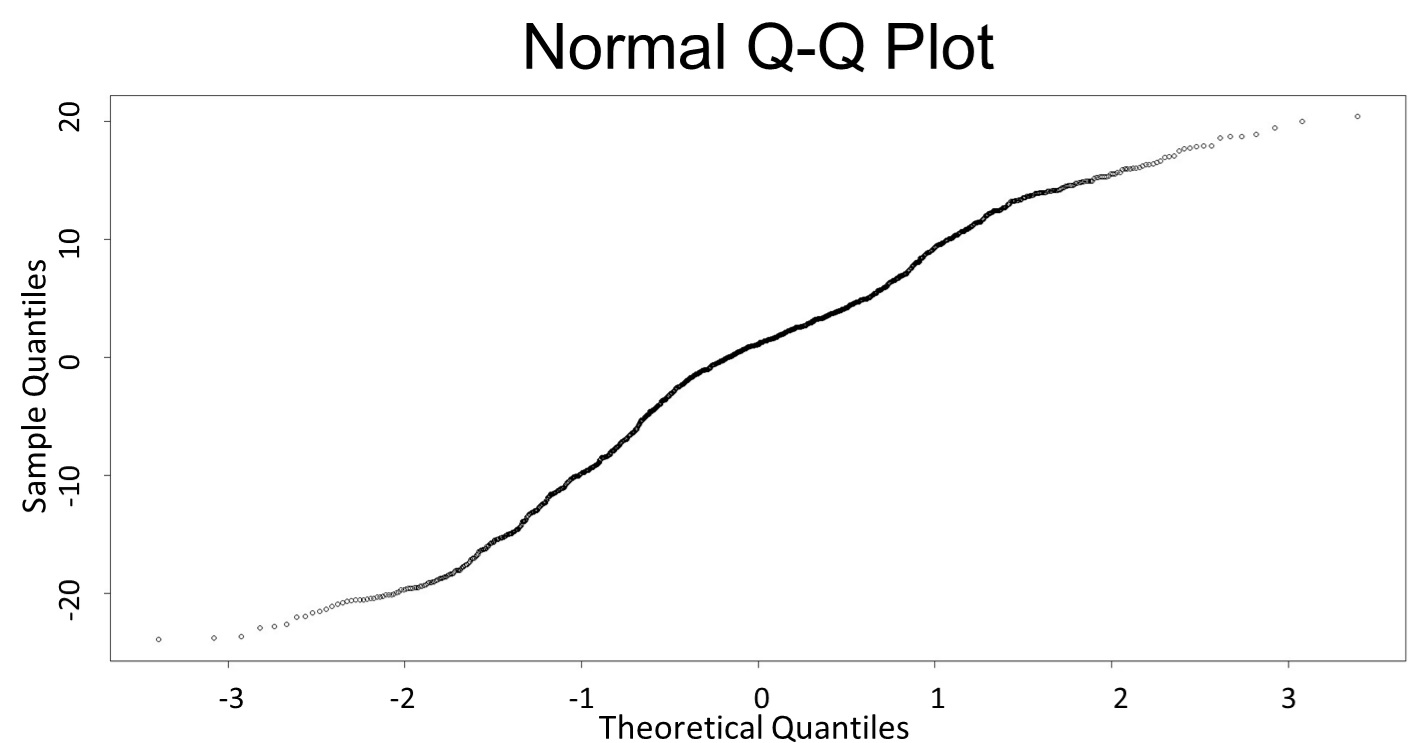
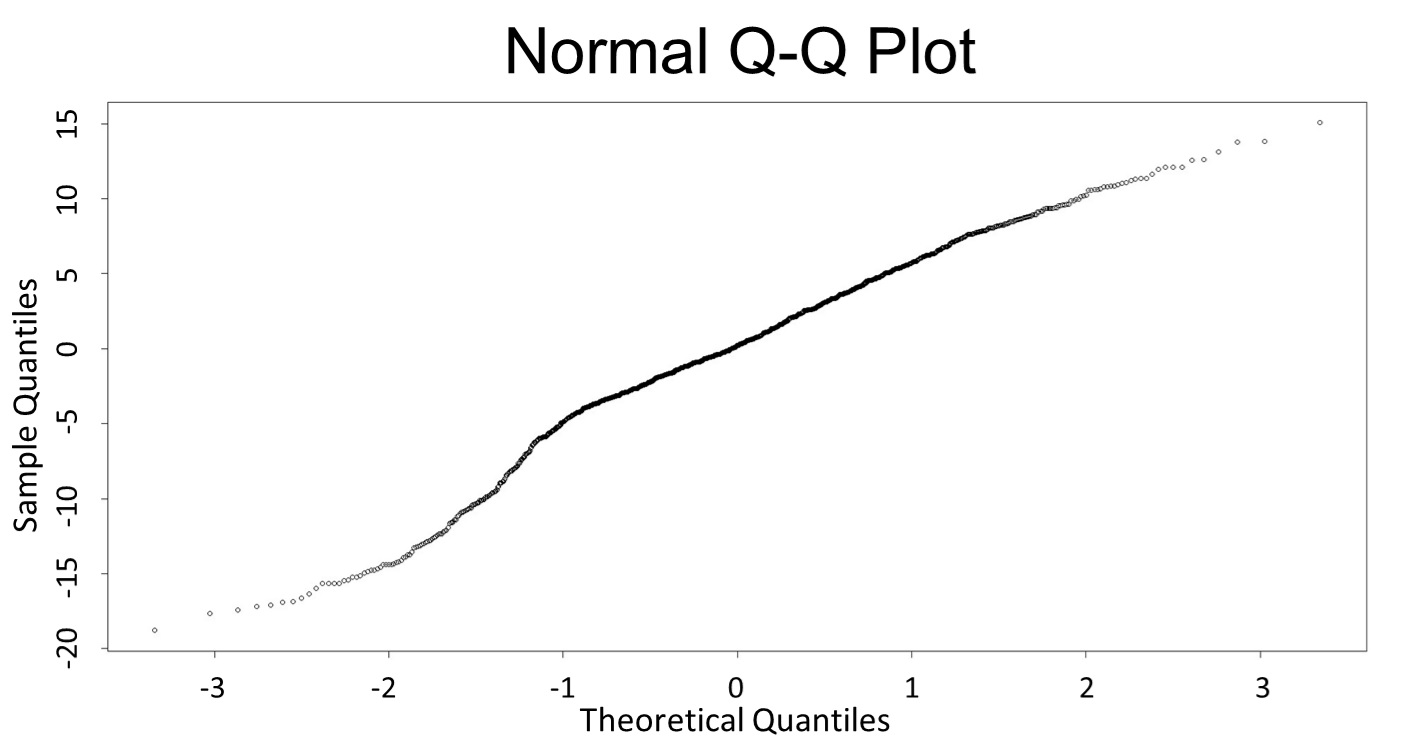
(Q-Q plot)
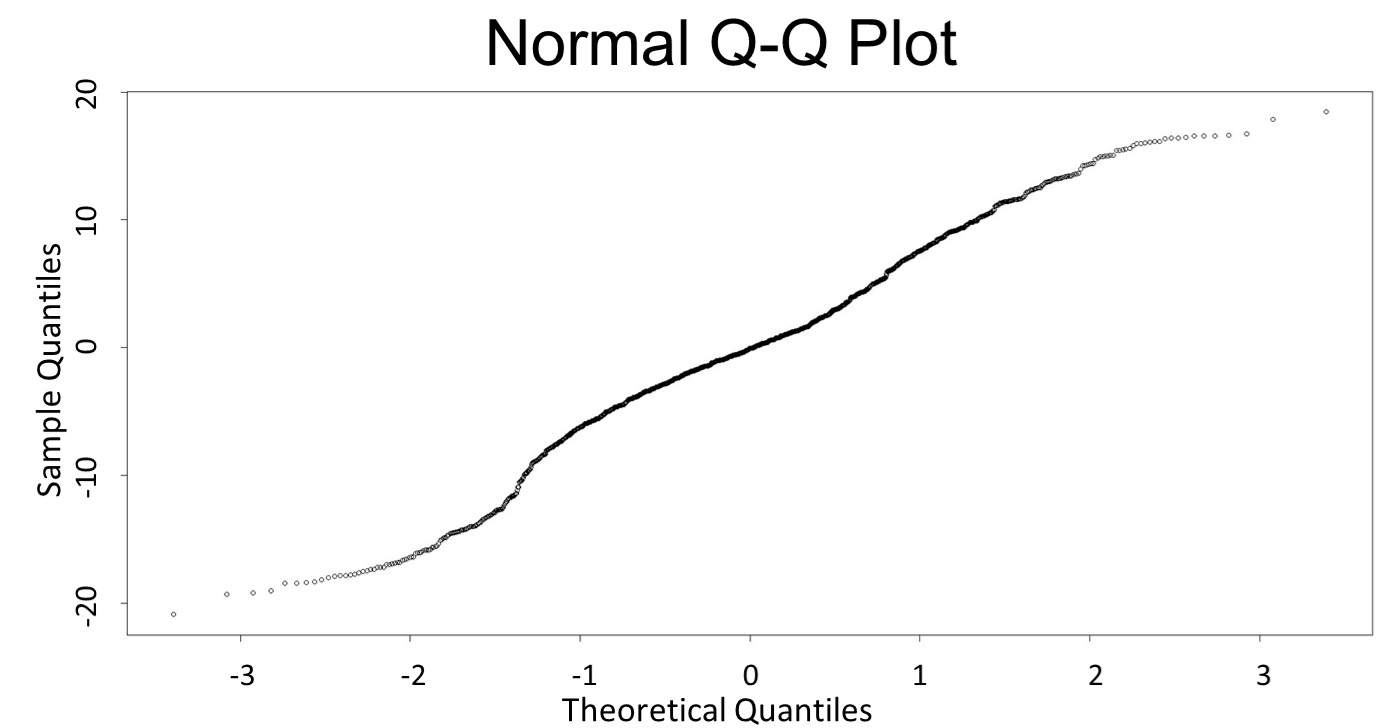
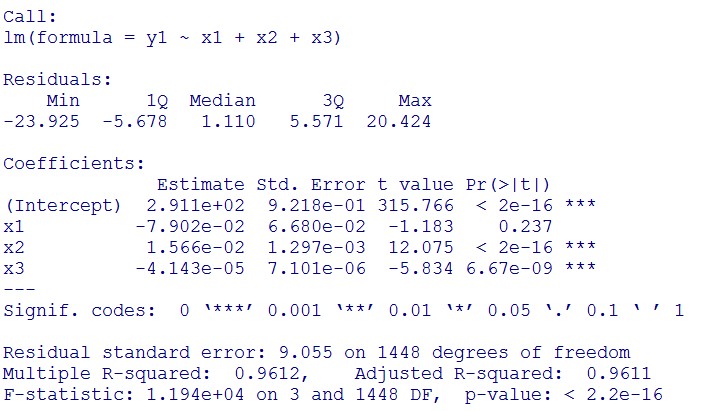
This model conforms the assumption and R-square is equal to 0.9541, so we think the cubic polynomial regression model is suitable.
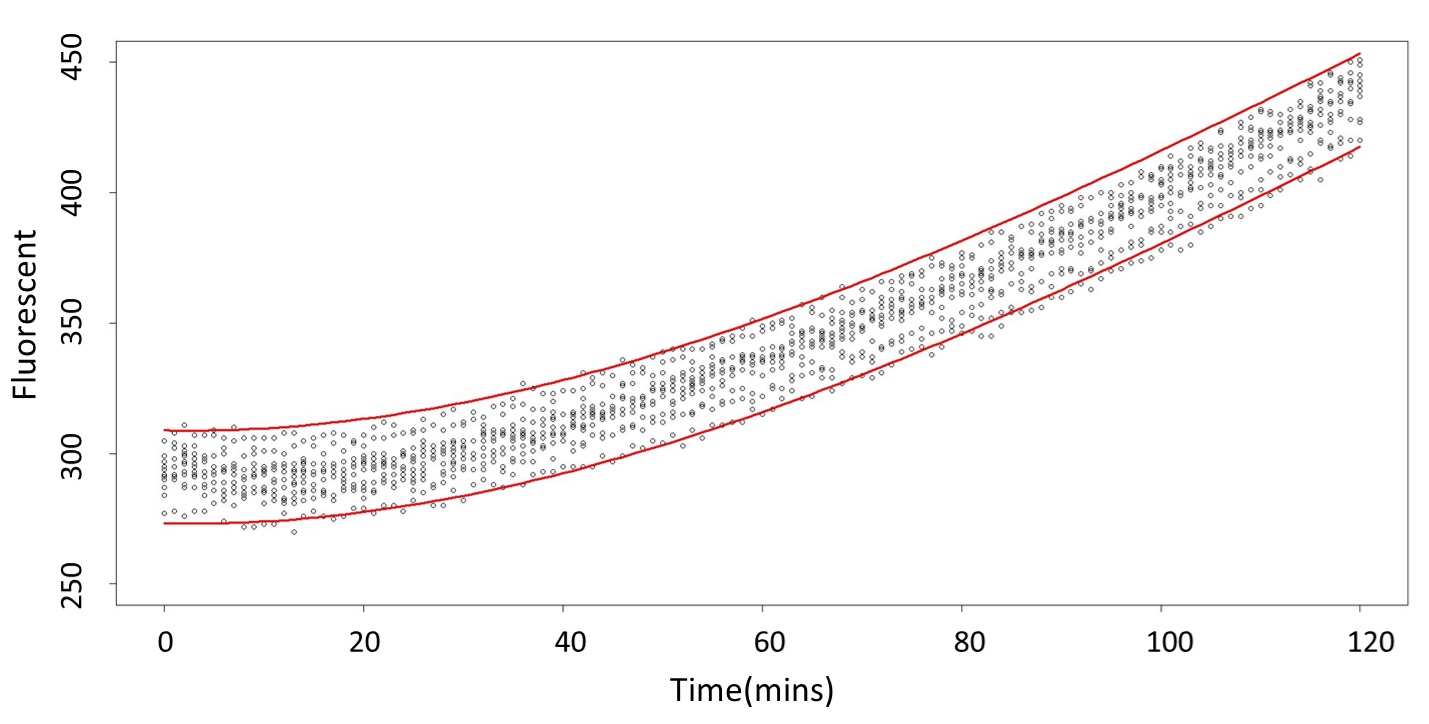
(prediction interval)
Formula: \(\bar{X}\pm t_{\frac{\alpha}{2},n-1}S\sqrt{1+\frac{1}{n}}\)
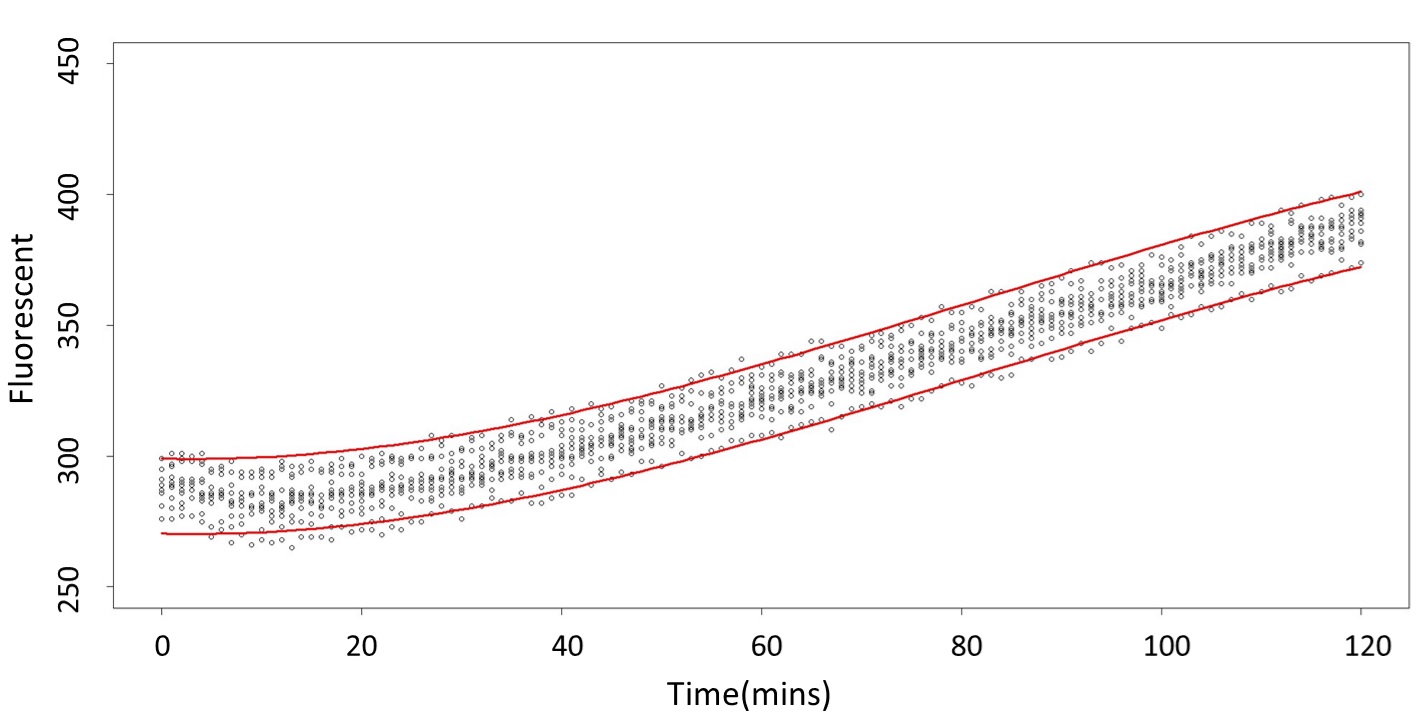
And the procedure is same in the 5mM and 120mM.
(5mM)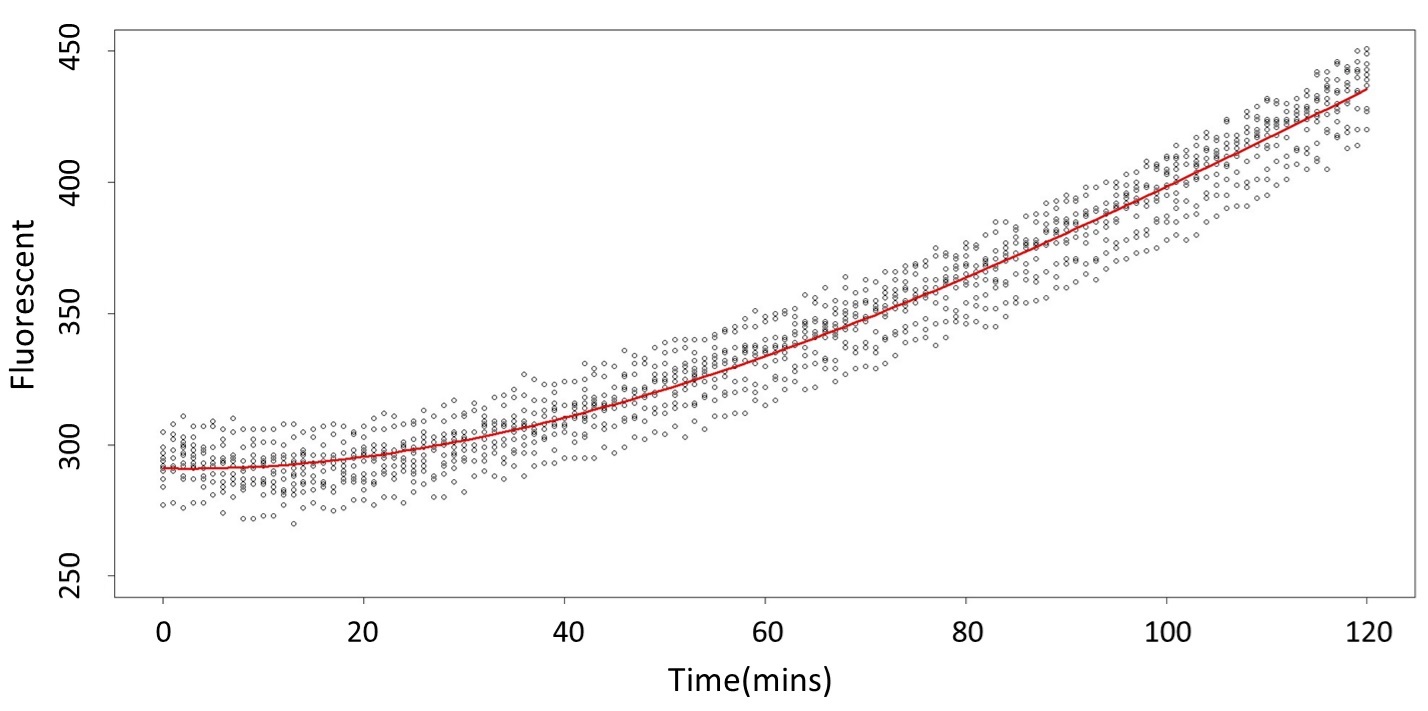
 (120mM)
(120mM)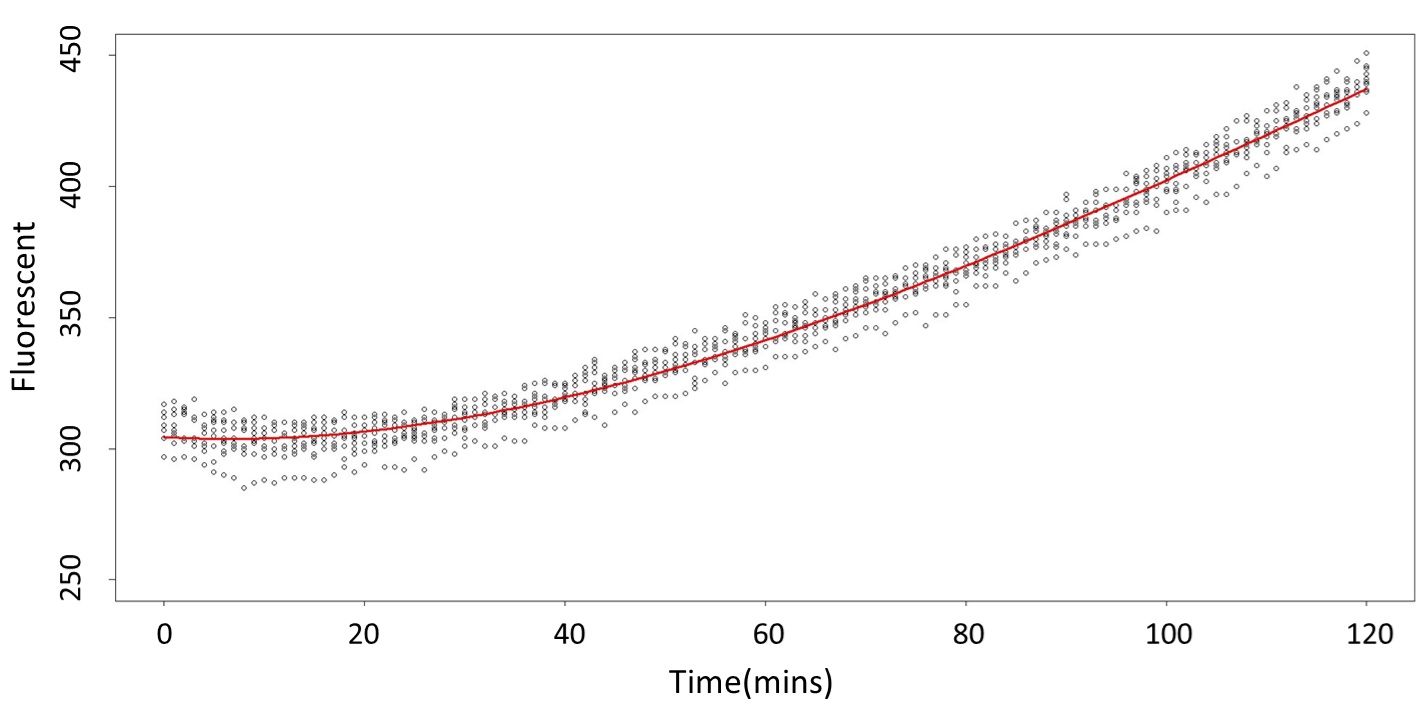
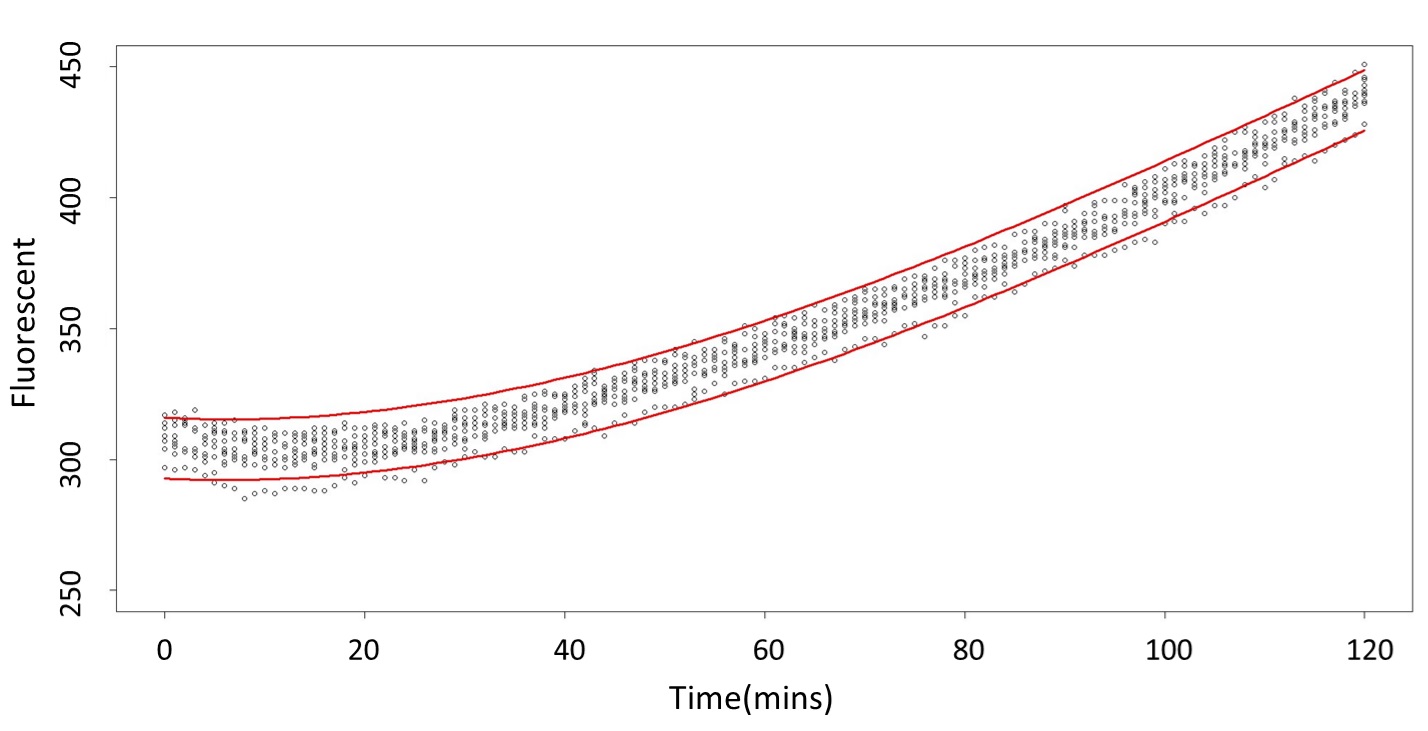
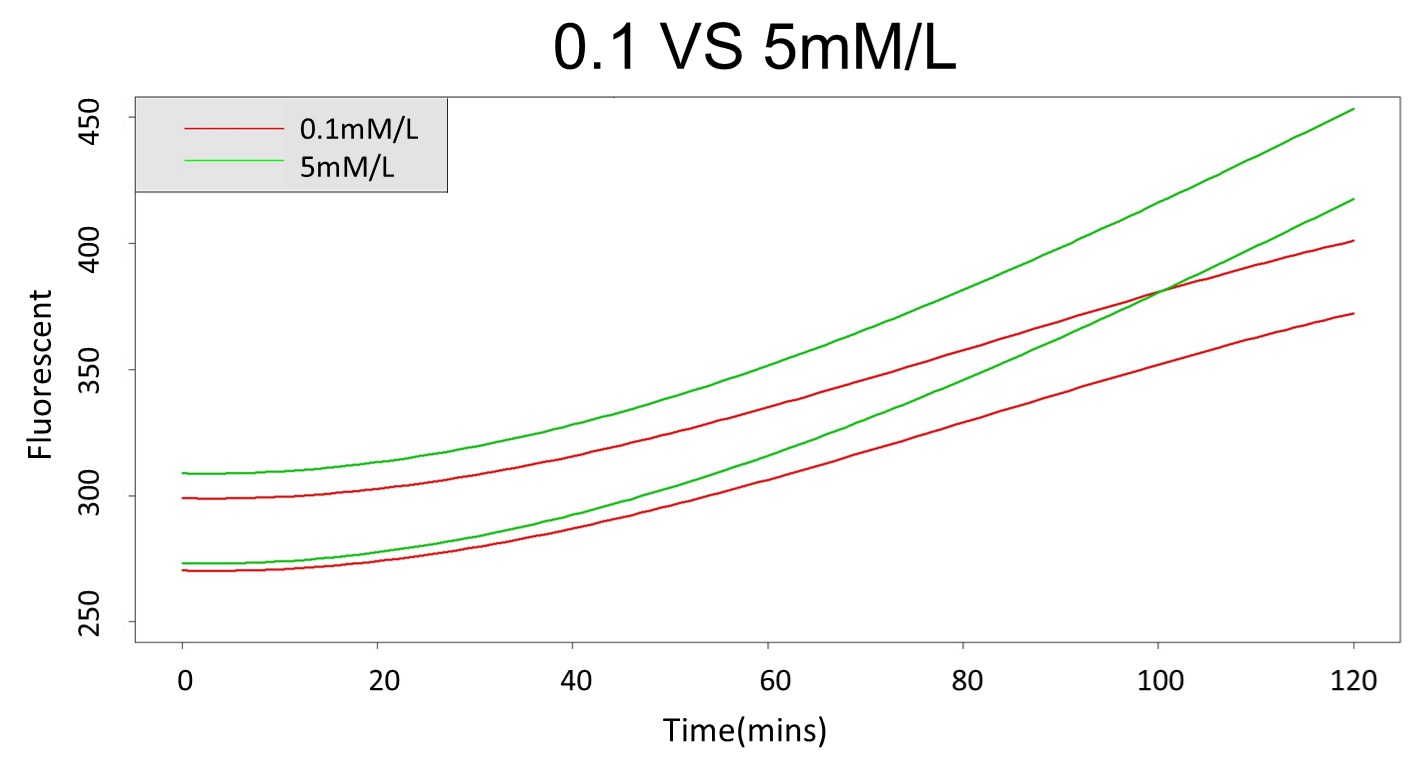
2. According to the paired-difference T test part, it demonstrates that 0.1mM and 5mM can be identified as different group due to the statistical significance. In order to precisely distinguish these two groups, choose the time at which the 5mmo/L and 0.1mmo/L prediction interval started to diverge to be the recommended prediction time.

Based on the above tables, it showed a significant degree of accuracy when an intersection upon the maximum fluorescence intensity at 0.1 mM and the minimum fluorescence intensity at 5 mM occurs after 101 mins (t> 101 mins). Hence, it can be expected to have higher accuracy of data after 101 mins.
(prediction interval of 0.1 &5 mM)

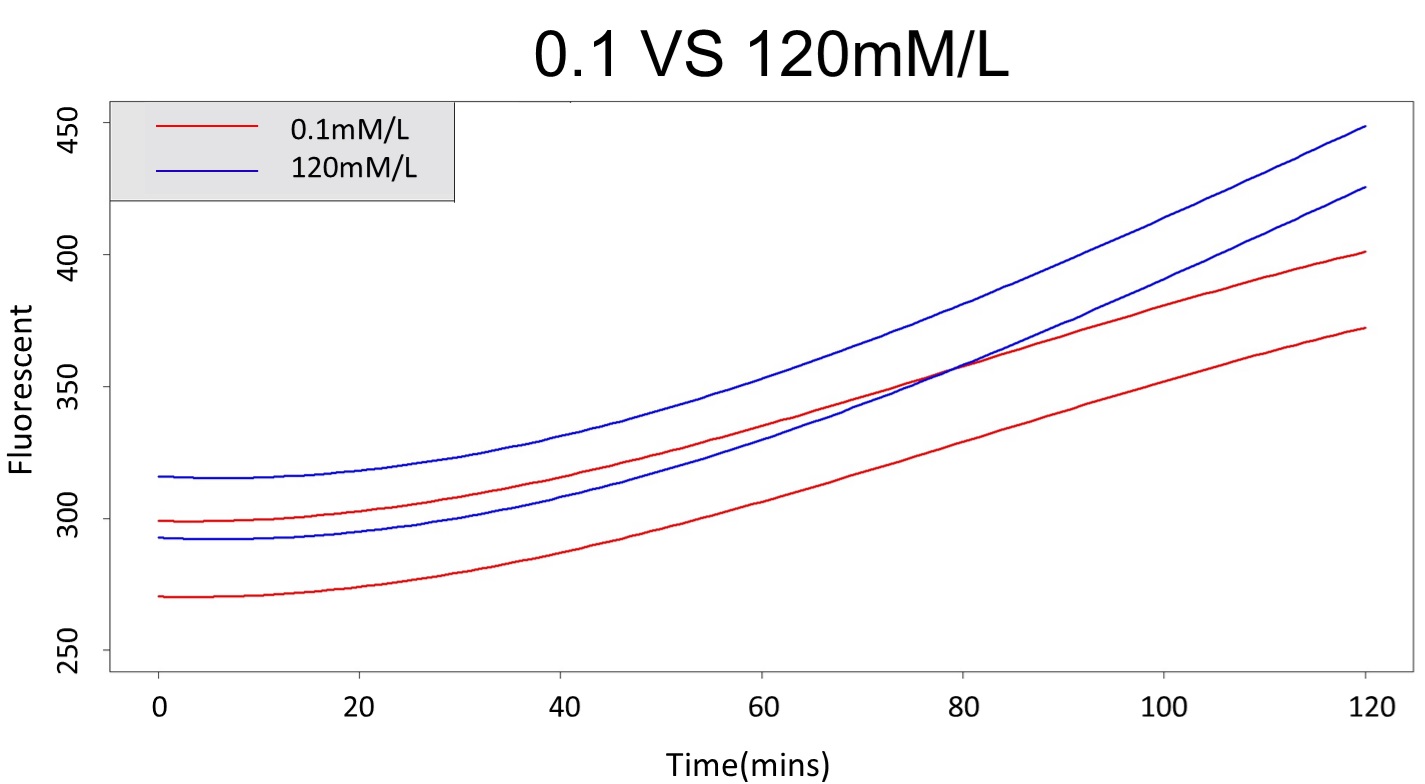
3. As same as Procedure 2, we choose the time at 120mmM and 0.1mM prediction interval to diverge to be the recommended prediction

According to the above tables, it showed a significant degree of accuracy when an intersection upon the maximum fluorescence intensity at 5 mM and minimum fluorescence intensity at 120 mM occurs after 88 mins (t> 88 mins). Hence, it can be expected to have higher accuracy of data after 88 mins.
(prediction interval of 0.1 &120 mM)

[1]MONTGOMERY D., et al., Introduction to linear regression analysis, Chapter 2-4 (2012)
[2]Mansfield ER., et al., Diagnostic value of residual and partial residual plots, Journal of The American Statistician (1987)
#0.1 vs 5
#0.1 urine plot
data<-read.csv("C:/Users/Rick/Desktop/NCKUactivity/iGEM/819test/0.1urine.csv",header=T)
x1<-rep(data[,1],12)
x2<-x1^2
x3<-x1^3
y1<-c(data[,2],data[,3],data[,4],data[,5],data[,6],data[,7], data[,8],data[,9],data[,10], data[,11],data[,12],data[,13])
model<-lm(y1~x1+x2+x3)
yhat0.1<-model$fit
plot(x1,y1,ylim=c(250,450),xlab= "time(mins) ",ylab= "Fluorescent", cex.lab=1.5)
#lines(data[,1], yhat0.1[1:121],lwd=3,col=2)
n=121*12
mse<-sum((y1-yhat0.1)^2)/(n-3) # calculate MS_Res(=σ^2)
X<-cbind(1,x1,x2,x3)
se<-sqrt(mse*(1+X[1,]%*%solve(t(X)%*%X)%*%X[1,]))
up0.1<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.975,df=n-2)*se
low0.1<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.025,df=n-2)*se
lines(data[,1], up0.1[1:121], col=2,lwd=3)
lines(data[,1], low0.1[1:121],col=2,lwd=3)
#5 urine plot
data<-read.csv("C:/Users/Rick/Desktop/NCKUactivity/iGEM/819test/5urine.csv",header=T)
x1<-rep(data[,1],12)
x2<-x1^2
x3<-x1^3
y1<-c(data[,2],data[,3],data[,4],data[,5],data[,6],data[,7], data[,8],data[,9],data[,10], data[,11],data[,12],data[,13])
model<-lm(y1~x1+x2+x3)
yhat5<-model$fit
par(new=TRUE)
plot(x1,y1,ylim=c(250,450),xlab= "time(mins) ",ylab= "Fluorescent", cex.lab=1.5)
#lines(data[,1], yhat5[1:121],lwd=3,col=2)
n=121*12
mse<-sum((y1-yhat5)^2)/(n-3) # calculate MS_Res(=σ^2)
X<-cbind(1,x1,x2,x3)
se<-sqrt(mse*(1+X[1,]%*%solve(t(X)%*%X)%*%X[1,]))
up5<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.975,df=n-2)*se
low5<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4] +qt(.025,df=n-2)*se
lines(data[,1], up5[1:121], col=3,lwd=3)
lines(data[,1], low5[1:121], col=3,lwd=3)
title(main="0.1 vs 5mM",cex.main=4)
Name<-c(expression(paste("0.1mM")),expression(paste("5mM")))
legend("topleft", Name, ncol = 1, cex = 1.5, col=c("red","green"),lwd = c(2,2), bg = 'gray90')
#0.1 vs 120
#0.1 urine plot
data<-read.csv("C:/Users/Rick/Desktop/NCKUactivity/iGEM/819test/0.1urine.csv",header=T)
x1<-rep(data[,1],12)
x2<-x1^2
x3<-x1^3
y1<-c(data[,2],data[,3],data[,4],data[,5],data[,6],data[,7], data[,8],data[,9],data[,10], data[,11],data[,12],data[,13])
model<-lm(y1~x1+x2+x3)
yhat0.1<-model$fit
plot(x1,y1,ylim=c(250,450),xlab= "time(mins) ",ylab= "Fluorescent", cex.lab=1.5)
#lines(data[,1], yhat0.1[1:121],lwd=3,col=2)
n=121*12
mse<-sum((y1-yhat0.1)^2)/(n-3) # calculate MS_Res(=σ^2)
X<-cbind(1,x1,x2,x3)
se<-sqrt(mse*(1+X[1,]%*%solve(t(X)%*%X)%*%X[1,]))
up0.1<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.975,df=n-2)*se
low0.1<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficiens[4]+qt(.025,df=n-2)*se
lines(data[,1], up0.1[1:121], col=2,lwd=3)
lines(data[,1], low0.1[1:121],col=2,lwd=3)
#120 urine plot
data<-read.csv("C:/Users/Rick/Desktop/NCKUactivity/iGEM/819test/120urine.csv",header=T)
x1<-rep(data[,1],10)
x2<-x1^2
x3<-x1^3
y1<-c(data[,2],data[,3],data[,4],data[,5],data[,6],data[,9],data[,10],data[,11],data[,12],data[,13])
model<-lm(y1~x1+x2+x3)
yhat120<-model$fit
par(new=TRUE)
plot(x1,y1,ylim=c(250,450),xlab= "time(mins) ",ylab= "Fluorescent", cex.lab=1.5)
#lines(data[,1], yhat120[1:121],lwd=3,col=2)
n=121*10
mse<-sum((y1-yhat120)^2)/(n-3) # calculate MS_Res(=σ^2)
X<-cbind(1,x1,x2,x3)
se<-sqrt(mse*(1+X[1,]%*%solve(t(X)%*%X)%*%X[1,]))
up120<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.975,df=n-2)*se
low120<-model$coefficients[1]+x1*model$coefficients[2]+x2*model$coefficients[3]+x3*model$coefficients[4]+qt(.025,df=n-2)*se
lines(data[,1], up120[1:121], col=4,lwd=3)
lines(data[,1], low120[1:121], col=4,lwd=3)
title(main="0.1 vs 120mM",cex.main=4)
Name<-c(expression(paste("0.1mM")),expression(paste("120mM")))
legend("topleft", Name, ncol = 1, cex = 1.5, col=c("red","green"),lwd = c(2,2), bg = 'gray90')
