SMITe - The Concept
In short
The Issue
- Chronic wounds and antibiotic resistance are issues increasing in importance and often go hand-in-hand
- Current treatment strategies for chronic wounds rely on mechanical removal of dead tissue, or systemic antibiotic therapy, which are largely ineffective or come with considerable risks for the patient
- A major reason for ineffective treatments are biofilms; structures secreted by bacteria to facilitate establishment of a chronic infection
Our goals for SMITe - Spider Silk Mediated Infection Treatment:
We aim to construct a functional dressing for chronic wounds, using spider silk as a scaffold to attach biofilm-degrading enzymes. This includes:
- Expressing biofilm-degrading 'combat proteins’ and quantifying their potency
- Assembling combat proteins with a linker and tag needed for purification and attachment to spider silk
- Attaching the enzymes to spider silk using Sortase A, a transpeptidase
- Isolating Sortase A from the S. aureus genome and creating a ‘Sortase Package’ including basic parts needed for other iGEM teams to use Sortase in their own projects
- Testing the efficacy of the fully assembled wound dressing with attached enzymes on biofilms
Our Path
A Project is Born
We are delighted to say that from its inception, our project has benefitted from being a truly shared endeavor; one which we cannot wait to show you.
Following initial selection of the team members, we undertook a series of small group exercises which merged team building with the generation of new ideas. Once these were narrowed down to several per group, we tested the ease with which each group could present an unknown idea to the rest of the team. This was a very revealing experiment in terms of the generalisability of an idea, as well as how marketable each one could be.
It became clear rapidly that the team were stuck between a focus on biomaterials and health, and it was alongside the opportunity to work with recombinant spider silk that our final project plan evolved, joining these two fields perfectly.
Chronic Wounds
Chronic wounds are a major source of disability and high cost for healthcare systems worldwide, not to mention pain, misery and the continual threat of infection for the affected individual. Referred to as a “snowballing threat to public health”, chronic wound sufferers have been aptly described as “carriers of a silent epidemic”, who are often already diagnosed with “highly branded” diseases including diabetes and obesity [1]. In the current context of these conditions twinned with an aging population, the trend in chronic wounds is set only to rise. We decided to approach this clinical problem using Staphylococcus aureus biofilm as a model. Since this organism is part of our skin flora, it benefits from easy access to broken skin and easily becomes established in non-healing wounds; it is suggested that S. aureus is present in more than 90% of all chronic wounds [2].
A Look Beyond
In addition, questions on a pressing global public health threat are raised by our project: antimicrobial resistance. It is well established that bacterial biofilms, fostered by the chronic wound environment, protect bacteria from both antibiotics and an individual’s immune system, generating the perfect backdrop for increasing drug resistance [1]. We could not fail to acknowledge the intrinsic link between this world issue and our project plan and as such made it the focus of our final Human Practices and Safety theme.
Biofilm
With both chronic wounds and the variety of bacterial resistance mechanisms in our minds, we were concerned about using traditional bactericidal approaches to aid chronic wound healing, which often stumble into one of two pitfalls:
- a largely unspecific effect which risks to further aggravate the wound [3]
- a limited efficacy on chronic wounds from the very start and / or decreasing efficacy during the course of the treatment [4]
So instead, iGEM Stockholm decided to focus on a prominent feature of many chronic wounds – the biofilm. Biofilms are complex extracellular structures generated by a variety of bacteria as a self-protection mechanism and are often increased in response to stress such as antibiotic treatment [5]. They are essential in supporting bacteria when they adopt a ‘sessile’ lifestyle in the form of chronic colonization; they interfere with inflammation meant to rid the wound of bacteria, help the bacteria attach and detach from the wound surface as the infection progresses, and have been found to contribute to quicker development of antibiotic resistance [6] [7]. In particular the biofilms' contribution to antimicrobial resistance is becoming increasingly problematic and inspired a large proportion of our work in Human Practices.
The components of biofilms differ among bacterial species, but many of them have been found to serve a specific purpose. Three structures observed commonly in different biofilms are: exopolysaccharides, extracellular DNA, and a variety of biofilm-associated proteins [8].
The vital role that biofilm plays in persistent infections provides a solid rationale for making them our primary target.
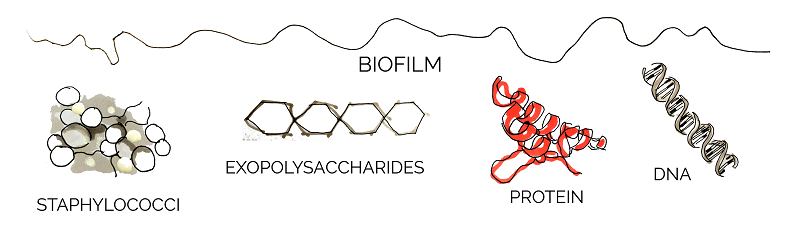
Our Project
Our vision is a functional wound dressing which will be able to disperse early-stage S. aureus biofilms in chronic wounds. The scaffold for this dressing is formed by spider silk – a material not only known for its high tensile strength, but also for being biocompatible and largely non-immunogenic. Those features make it an ideal candidate material for potential wound dressings [9]. Since the synthesis of spider silk itself is extremely complex and would constitute a project of its own, we were very lucky to receive ready-made, recombinant spider silk from our adivsors.
To actively fight the biofilm, several biofilm-degrading 'combat proteins' will be attached to the silk via Sortase A, a transpeptidase isolated from S. aureus. This constitutes a broadly-based approach which will hopefully forestall any bacterial escape or resistance mechanisms.

However, our mission is far from being limited to chronic wound treatment – products similar to our wound dressing could find applications far beyond the medical field.
Combat Proteins
eDNA and Nuclease
Key Achievements
Assembly and expression
Demonstrating dispersal of P. aeruginosa and S. aureus biofilm
Extracellular DNA (eDNA) is released by bacteria through autolysis (self-degradation) and serves as a matrix component of biofilm. eDNA is involved in various biofilm processes including adhesion, aggregation of bacterial cells and as a nutrient source. Enzymatic degradation of eDNA can weaken the biofilm and make the bacteria more susceptible to antibiotics and biocides [10]. Numerous studies have shown that biofilm-degradation can be achieved by a variety of DNases, enzymes that cleave single-stranded and double-stranded DNA [11]. In our project we aimed to degrade biofilm formed by S. aureus which has previously been shown to be susceptible to DNase treatment [12]. The DNase we used in the scope of this project was a nuclease (Nuc).
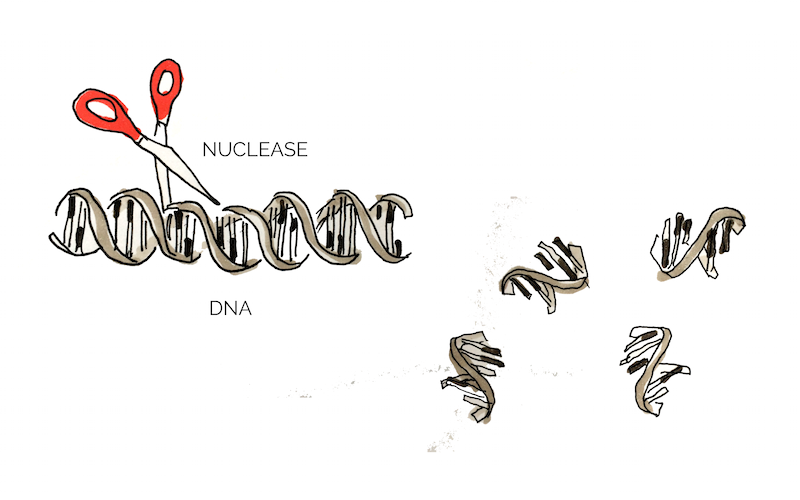
In addition to eDNA, nuclease may also cleave neutrophil extracellular traps (NETs) which are secreted by white blood cells in wounds as a response to bacterial infection [10]. Thus nucleases may interact negatively with the body’s own immune system and it would be of great advantage to create a nuclease that targets bacterial, but not human DNA. Eukaryotic and prokaryotic DNA typically exhibit different methylation pattern and some studies have taken advantage of this difference to specifically target bacterial DNA using methylation-dependent nucleases [13].
To target biofilm, we ordered a nuclease BioBrick BBa_K729004. Together with a T7 promoter BBa_I719005, we aimed to express Nuc, characterise its ability to disperse and inhibit biofilms formed by different bacteria and find out whether it displayed any kind of specificity for procaryotic or eucaryotic DNA. Whereas no clear inhibition of biofilm formation was shown, indications for successful dispersal were found.
Exopolysaccharides and Lysostaphin
Key Achievements
Assembly and expression
Testing of bactericidal activity on E. coli
Fulfilling a silver requirement: Demonstrating that lysostpahin works as expected by demonstrating its bacteriolytic activity and its ability to inhibit and disperse S. aureus biofilms
After initial formation, when the biofilm enters the maturation phase of development, intercellular aggregation initiates the formation of the 3D biofilm structure, characterized by cell ‘towers’ equipped with fluid-filled channels. In staphylococci, these structures are predominantly supported by an intricate network of beta-1,6-linked N-acetylglucosamine (PNAG) and other polymers including teichoic acids. These exopolymers are partially de-acetylated which generates a positive charge on the network. Since cell surfaces are naturally negative in charge, the electrostatic interaction between the cells and the exopolymers reinforces the build-up of the biofilm structure [14].
A strategy to disrupt the formation of biofilm would be to eradicate the production of S. aureus mediated PNAG exopolymers. Hence, to perform this particular action we chose lysostaphin, a glycyl-glycine endopeptidase, as one of our combat proteins.
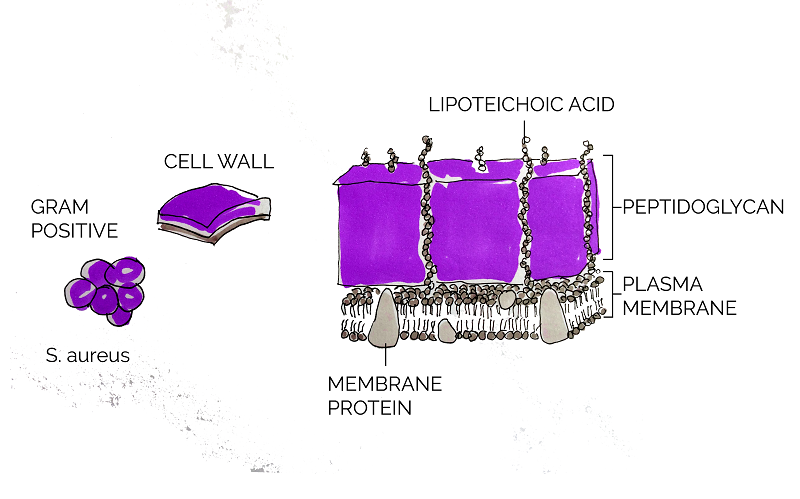
As a Gram positive bacterium, Staphylococcus aureus is encapsulated by a thick layer of peptidoglycan. Lysostaphin works by degrading this peptidoglycan layer which in turn results in cell lysis and death [15]. According to Sabala et al [16], lysostaphin achieves this by cleaving the pentaglycine bridge cross-linking the parallel stems of the peptidoglycan polymers.
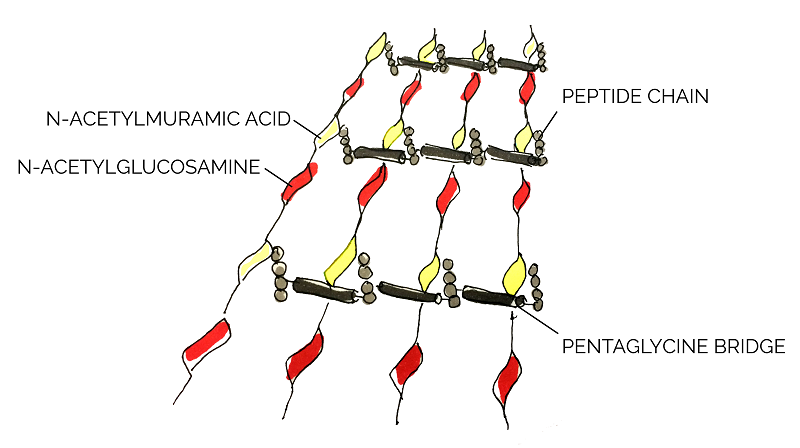
In order to manipulate and express lysostaphin in this project, we used an existing BioBrick, BBa_K748002. This truncated and mature form of lysostaphin was first developed by Team HIT-Harbin in 2012. With its specific-target mechanism as well as its relatively small size, lysostaphin can potentially be conjugated to spider silk as a combat protein to treat S. aureus infected wounds.
During the course of our project, we have achieved several of our goals with lysostaphin; after demonstrating expression, we started investigations into its solubility and bacteriolytic action and most importantly were able to show that lysostaphin strongly and specifically inhibited and degraded S. aureus biofilm.
Proteins and Esp
Key Achievements
Assembly and expression
Investigation into bactericidal activity on E. coli
Confirming Esp's ability to disperse biofilms
Fulfilling a gold requirement: Improving the characterisation of a previous team's Esp BioBrick in a new expression system and expand on their finding regarding biofilm dispersal and inhibition.
Proteins in the biofilm matrix have various functions and play an important role in all stages of the biofilm development. Cell wall anchored (CWA) proteins are vital in Staphylococcus aureus biofilms to mediate the primary attachment of cells to the surface of its host and also promote the intercellular adhesion, biofilm accumulation and maturation [17].
Staphylococcus epidermidis is a commensal species with S. aureus, and those two species competes with one another for nutrients. Epidemiology studies show that S. epidermidis inhibit the growth of S. aureus in the human nasal cavity and further studies indicated that this was due to the degradation of a variety of S. aureus CWA proteins. This degradation resulted in decreased biofilm formation and forced S. aureus back into a planktonic form, presumably resulting in decreased survival chances. [18] [19] Heavily involved in this process was extracellular serine protease (Esp); secreted by S. epidermidis, this protease degrades the proteins that maintain structural and chemical integrity of the biofilm. Purified Esp inhibits biofilm formation at the preliminary stage and is able to detach a pre-existing one. Additionally, Esp reduces the virulence of S. aureus through degrading other important proteins, e.g. quorum sensing molecule receptors, enzymatic proteins for biofilm dispersal. [20]
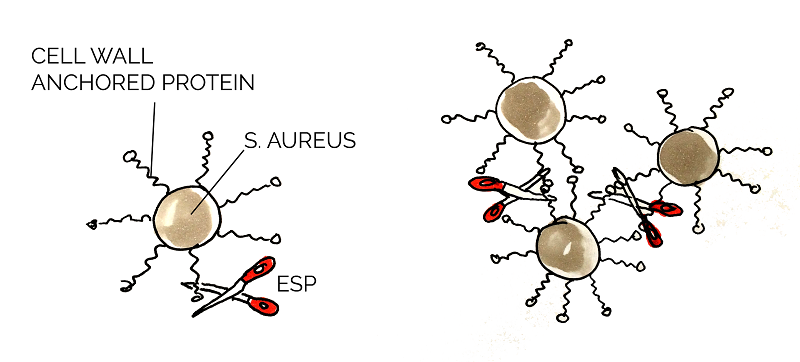
Esp could be applicable for the prevention or treatment of infectious diseases caused by S. aureus. It has also been proven to be effective for the biofilms formed by MRSA and vancomycin-intermediate S. aureus. [19] We used the Biobricks BBa_K531003 and BBa_K531006, which were developed by team Grinnell 2011.
Esp is produced as an inactive form of protein which is then secreted into the environment through Sec-mediated translocation. The propeptide at the N-terminal would be cleaved by signal peptidases during or right after the protein secretion. When expressed recombinantly, the protein therefore requires enzymatic activation - an issue we chose to address by working to create a truncated, mature version of the protein. [21] A successfully expressed Esp could be a good choice to tackle S. aureus biofilm through multiple pathways.
We aimed to improve the characterization of the Esp BioBrick by (1) testing expression in a new chassis, E. coli, (2) validating the findings of the creators indicating that Esp was able to disperse S. aureus biofilm and expand on this by testing other bacterial biofilms. To our knowledge, we were also the first iGEM team to characterize this BioBrick’s ability to (3) inhibit biofilm formation and (4) test a potential bacteriolytic activity.
We found that expression in E. coli was not very strong and Esp failed to show bacteriolytic activity or clear inhibition of biofilm formation by S. aureus and P. aeruginosa in our setup. However, Esp was able to significantly detach established biofilms of both species. You can read more about our findings on the results page on the registry.
Cell membranes and Defensin
Key Achievements
Assembly and expression
Testing activity on S. aureus and P. aeruginosa biofilm
Addition of LT
Expression and purification using IMAC
Conjugation to Spider Silk
Defensin is an antimicrobial peptide (AMP) that contributes to the innate immune response and is present in both vertebrates and invertebrates. It is highly abundant in tissues involved in the host defense system and its amphipathic character allows it to penetrate and disrupt the integrity of pathogenic microbial membranes for destruction. Honey-bee (Apis mellifera) derived defensin-1 was shown to successfully reduce the viability of biofilm-encapsulated wound pathogens, including P. aeruginosa and our main target, S. aureus [22].
The function of defensin is to lyse the pathogenic cell by permeabilizing the plasma membrane. The surface of defensin has both hydrophobic and hydrophilic regions. Upon binding to the microbial membrane, defensin causes the formation of small channels, disrupting the integrity and increasing permeability of the membrane. This results in efflux (outward movement from cell) of vital components in the cell, thus killing it [23].
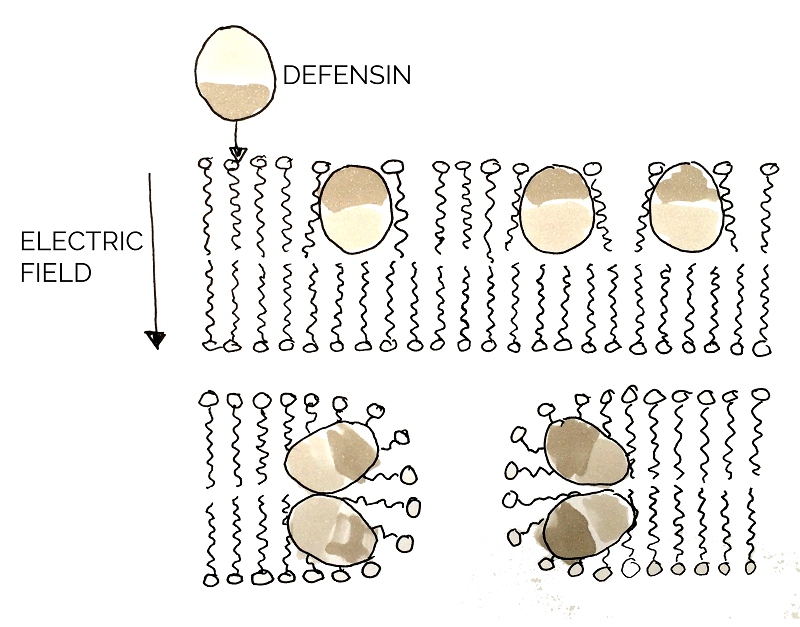
As our primary target in chronic wounds is the infectious bacteria S. aureus and considering that defensin has successfully been shown to have a negative effect on membranes of pathogenic organisms, [24] defensin was considered a great combat protein candidate. The well-studied, honeybee-derived defensin-1 part BBa_K1104301, from the Bee.coli project designed by 2013 NYMU iGEM Team-Taipei, was ordered for usage in SMITe and expressed. A successfully produced and functional defensin would optimize the disintegration of several bacterial components of the chronic wound model by killing S. aureus, amongst other pathogens in the biofilm structure.
Defensin proved to be one of our most successful proteins in the lab - we were able to demonstrate expression of defensin as well as a strong biofilm-inhibiting activity. Additionally, we assembled a defensin construct which allowed us to purify and finally conjugate the protein to spider silk - a big step for the development of SMITe.
Sortase A
The Enzyme
When designing the assembly of the essential parts SMITE constitutes of, i.e. the silk and the combat proteins, we were aiming to create an easily-applicable system for attaching proteins to the silk. The main concern was to be able to attach proteins to the silk without altering their main structure, in fear of disturbing the integrity of the protein's structure and its functionality.
Additionally, the spider silk used in our project was produced by Spiber Technologies using a trademarked recombinant protein which meant we were not able to use a classical genetic engineering approach to create fusion proteins due to confidentiality. When looking for options to circumvent this issue, we came across Sortase A, a bacterial transpeptidase commonly expressed by Gram positive bacteria. It has the ability to break and form new peptide bonds through a nucleophilic attack and is used by Staphylococcus aureus to anchor virulence factors to the cell wall [25].
The key feature of the enzyme is the specific conjugation reaction it carries out, where the enzyme recognizes a specific amino acid sequence, a so called sorting motif (LPXTG motif in the case of S. aureus) [25] [26].
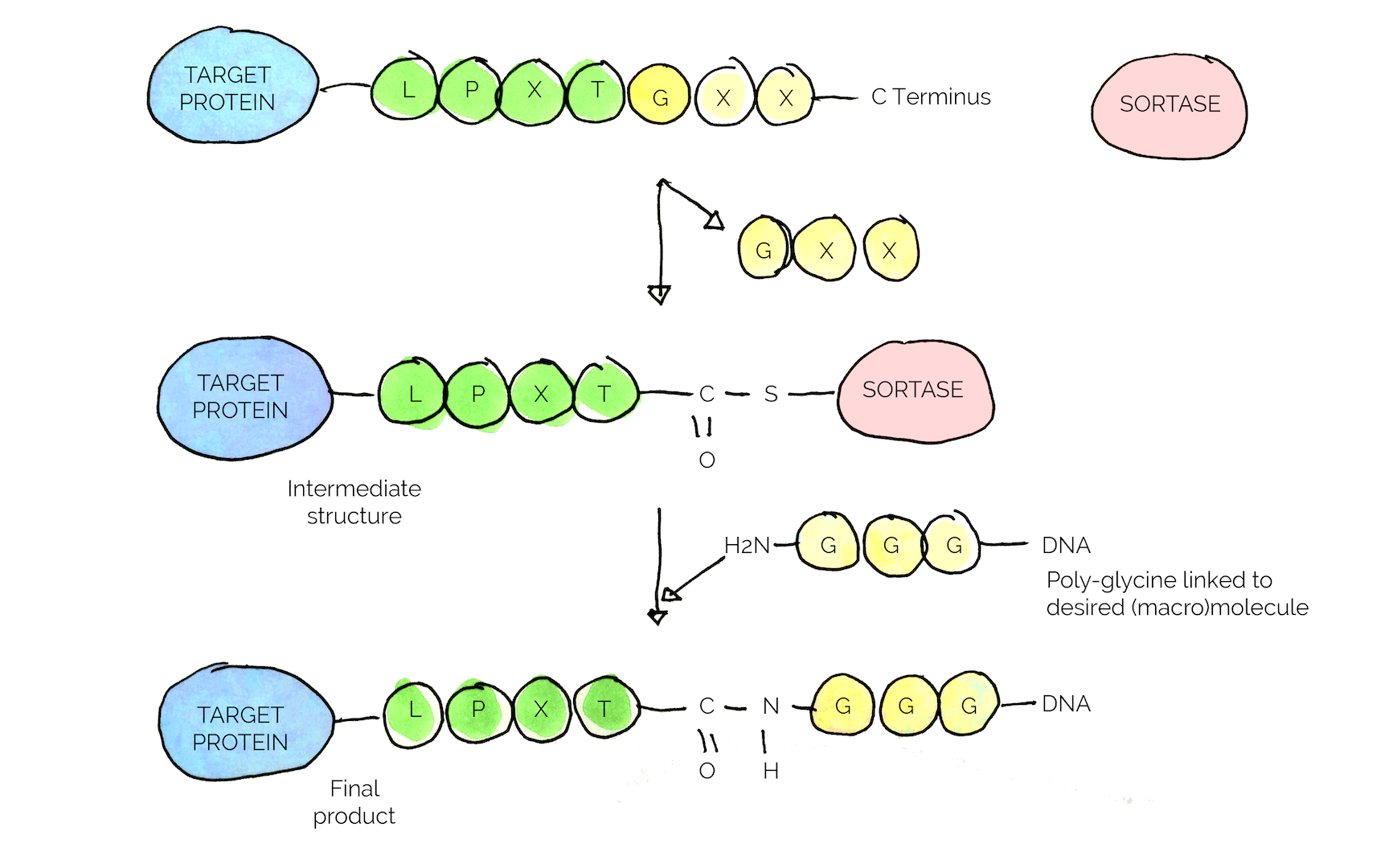
In our project, we chose to isolate the enzyme from S. aureus and make it into a standard BioBrick with an added GB1 sequence to improve solubility [26].
Our Sortase-A-mediated Immobilization Strategy
Key Achievements
Isolation of Sortase A from S. aureus genome
Assembly with solubility tag GB1
Successful conjugation of peptides to spider silk using commercial enzyme
We aimed to use the specific interaction of Sortase with the sorting motif to conjugate the “combat proteins” to the spider silk. A general strategy for all combat proteins was designed, in which a specific linker-tag sequence (LT) including the sorting motif and a His-tag for purification purposes, are fused with the proteins (see Figure 8), resulting in a finalized gene construct. Each protein when expressed will be equipped with the LT, enabling Sortase mediated conjugation/immobilization of the proteins to the spider silk naturally occurring C-terminal glycine.
During our iGEM project, we successfully isolated Sortase A from the S. aureus genome and employed two cloning strategies to achieve the fusion with GB1. Unfortunately, the problematic assembly meant that we were unable to assemble the full expression construct or conduct functional testing.
The Sortase Package
Sortase has been identified as an enzyme with a broad range of applications that can be extrapolated to the usage of the enzyme in future iGEM project serving as a potential standard BioBrick used within the iGEM Community.
We thus aimed to generalize our Sortase strategy further, to develop a system in which future iGEM teams can easily utilize Sortase to conjugate/immobilize any type of fusion partners of interest. For more information please see the Part Collection page.
References
1: Sen, C.K., Gordillo, G.M., Roy, S. et al. Human Skin Wounds: A Major and Snowballing Threat to Public Health and the Economy. Wound Repair Regen. 17(6):763-771 (2009)
2: Gjodsbol, K., Christensen, J.J., Karlsmark, T. et al. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J. 3(3):225-231 (2006)
3: Atiyeh, B. S., Dibo, S. A. & Hayek, S. N. Wound cleansing, topical antiseptics and wound healing. Int. Wound J. 6, 420–30 (2009)
4: Zhao, G. et al. Biofilms and Inflammation in Chronic Wounds. Adv. wound care 2, 389–399 (2013).
5: O’Toole, G. A. & Stewart, P. S. Biofilms strike back. Nat. Biotechnol. 23, 1378–1379 (2005).
6: Spiliopoulou, A. I. et al. Bacterial adhesion, intracellular survival and cytokine induction upon stimulation of mononuclear cells with planktonic or biofilm phase Staphylococcus epidermidis. FEMS Microbiol. Lett. 330, 56–65 (2012),
7: Stewart, P. S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292, 107–13 (2002)
8: Flemming, H.-C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–33 (2010)
9: Meinel, L. et al. The inflammatory responses to silk films in vitro and in vivo. Biomaterials 26, 147–55 (2005).
10: Okshevsky M, Regina VR, Meyer RL. Extracellular DNA as a target for biofilm control. Curr Opin Biotechnol. 2015 Jun;33:73-80.
11: Chen WJ1, Liao TH. Structure and function of bovine pancreatic deoxyribonuclease I. Protein Pept Lett. 2006;13(5):447-53.
12: Izano EA1, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008 Jan;74(2):470-6.
13: Siwek W1, Czapinska H, Bochtler M, Bujnicki JM, Skowronek K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restrictionendonuclease R.DpnI. Nucleic Acids Res. 2012 Aug;40(15):7563-72.
14: Otto, M. Staphylococcal Biofilms. Curr Top Microbiol Immunol. 322, 207-228 (2008)
15: Kumar, J. K. Lysostaphin: an antistaphylococcal agent. Appl Microbiol Biotechnol. 80, 555-561 (2008)
16: Sabala, I., Jagielska, E., Bardelang, P. T., Czapinska, H., Dahms, S. O., Sharpe, J. A., James, R., Than, M. E., Thomas, N. R. & M. Bochtler. Crystal structure of antimicrobial peptidase lysostaphin from Staphylococcus simulans FEBS Journal. 281, 4112-4122 (2014).
17: Pietro Speziale, Giampiero Pietrocola, Timothy J. Foster and Joan A. Geoghegan, Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol. 2014; 4: 171.
18: Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346 –349.
19: Sugimoto S, Iwamoto T, Takada K, Okuda K, Tajima A, Iwase T, Mizunoe Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol. 2013 Apr;195(8):1645-55.
20: Chen Chen, Vengadesan Krishnan, Kevin Macon, Kartik Manne. Secreted Proteases Control Autolysin-mediated Biofilm Growth of Staphylococcus aureus. J Biol Chem. 2013 Oct 11; 288(41): 29440–29452.
21: S. Sugimoto, T. Iwase, F. Sato, A. Tajima, H. Shinji and Y. Mizunoe. Cloning, expression and purification of extracellular serine protease Esp, a biofilm-degrading enzyme, from Staphylococcus epidermidis. J Appl Microbiol. 2011 Dec;111(6):1406-15.
22: Sojka M, Valachova I, Bucekova M, Majtan J. Antibiofilm efficacy of honey and bee-derived defensin-1 on multi-species wound biofilm. J Med Microbiol. April 2016 65: 337-344
23: Tomas Ganz. Defensins: antimicrobial peptides of innate immunity. Nature Reviews Immunology 3, 710-720 (September 2003)
24: M Shimoda, K Ohki, Y Shimamoto and O Kohashi. Morphology of defensin-treated Staphylococcus aureus. Infect Immun. 1995 Aug; 63(8): 2886–2891.
25: Popp, M. W.-L. and Ploegh, H. L. (2011), Making and Breaking Peptide Bonds: Protein Engineering Using Sortase. Angew. Chem. Int. Ed., 50: 5024–5032
26: Westerlund, K. Karlstrom, A., Honarvar, H., Tolmachev, V. Design, Preparation, and Characterization of PNA-Based Hybridization Probes for Affibody-Molecule-Mediated Pretargeting. Bioconjugate Chem., 2015, 26 (8), pp 1724–1736


