(→How Stable is Biotin Coupled to Proteins?) |
(→Stabilizing the Biotin Peptide Bond) |
||
| Line 48: | Line 48: | ||
===Stabilizing the Biotin Peptide Bond=== | ===Stabilizing the Biotin Peptide Bond=== | ||
| + | If we are to prevent biotinidase from cleaving biotin off of biotinylated components in our bioprinting system and thus from disintegrating our mesh while simultaneously obviating use of special serum-free media, we must consider how to render the biotin bound to the spacer peptide non-hydrolyzable by biotinidase.<br> | ||
| + | Biotinidase-resistant biotin conjugates have been extensively studied in the context of the cancer treatment "pretargeting" approach. Here, biotinylated IgG is bound to the tumor followed by sequential treatment with streptavidin and radionuclide-containing biotin, which offers the advantages of fast accretion as well as clearance combined with low tissue retention (Bogusiewicz et al., 2004; Goldenberg, Chatal, Barbet, Boerman, & Sharkey, 2007).<ref name=bogusiewicz2004/><ref name=goldenberg2007>Goldenberg, D. M., Chatal, J.-F., Barbet, J., Boerman, O., & Sharkey, R. M. (2007). Cancer Imaging and Therapy with Bispecific Antibody Pretargeting. Update on cancer therapeutics, 2(1), 19-31.</ref> The general consensus is that steric hindrance at and adjacent to the peptide bond to be cleaved is paramount; for instance, ''p''-aminobenzoate is cleaved off much faster than the ''m''-isomer, and the ''o''-isomer is not cleaved at all. Thus, resistance is acquired either if the amide nitrogen is tertiary, e.g. carrying an additional methyl group (as in biotinyl-sarcosine), or if the position ''next to'' this nitrogen carries a substituent.<ref name=chauhan1986/> Among the reported non-hydrolyzable biotin derivatives are those with the following substituents adjacent to the amide nitrogen: hydroxymethyl (serine), carboxylate (aspartate), ethyl (2-aminobutyric acid), isopropyl (valine), benzyl (phenylalanine); a methyl group (alanine), however, is not sufficient.<ref name=wilbur2006/><ref name=wolf1990/> | ||
| + | Since increased steric bulk diminishes the affinity of streptavidin toward the biotin, the 2-aminobutyric acid derivative has been preferred over e.g. the valine conjugate because it binds to streptavidin more strongly and is less lipophilic. However, this deviation may not ultimately be relevant given the inherent exceptionally high avidity of streptavidin toward biotin.<ref name=wilbur2006/> | ||
=Design= | =Design= | ||
Revision as of 21:26, 10 October 2016
Kopiervorlagen
Seitenverantwortliche/r:Vivien
Bei Google Scholar bitte das APA-Ziteirformat verwenden.
Textformatierung
kursiv
fett
Strich
Links
Wikiinterner Link Team:LMU-TUM_Munich/Materials (As described in the Materials section)
Wikiexterner Link Visit W3Schools
Visit W3Schools
Bilder
blablabla
blablabla
Introduction
Purpose
Any cell mesh produced based on our bioink will suffer from exposition to serum-containing media or, in the case of in vivo application, the patient’s body fluids because of biotinidase. This ubiquitous enzyme is able to cleave biotin off of lysine – at a lower rate also off of intact protein. We may now shift this hydrolysis in two directions: either to prevent it altogether and thus stabilize our cell mesh, or to accelerate it to make our mesh quickly disintegrable. To this end, we must consider how biotin is linked to the proteins in our bioink.
NHS Esters as Amine Coupling Reagents
N-hydroxysuccinimide (NHS) esters belong to the group of amine-reactive active esters and are thus of great use in labeling proteins. Typically, NHS esters react with the primary amino group in the side chain of surficial lysine, forming a stable peptide bond.[1]
The utility of NHS esters was first suggested by Anderson et al.[2] in the context of peptide synthesis; nowadays, a plethora of NHS esters are widely used in bioconjugate techniques and thus commercially available, including those of many fluorescent dyes as well as that of biotin, often referred to as biotin-NHS (Figure 1).[3] NHS esters are typically synthesized from the corresponding carboxylic ester and NHS via the one-pot DCC coupling procedure[1] and purified by crystallization[2][3].
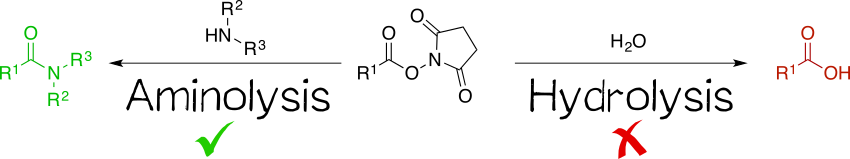
Being amine reactive, NHS esters undergo an SN2t reaction with primary or secondary amines to afford an amide bond as well as free NHS (Scheme 1, left). While this aminolysis pathway is also used in chemical synthesis, e.g. of peptides, it is key for the above-mentioned protein-labeling capability of NHS esters. The latter, however, strictly requires aqueous reaction media, so that the undesirable hydrolysis, i.e. reaction with water to give the no longer reactive carboxylic acid (Scheme 1, right), becomes a considerable side reaction even though reaction with amines, being more nucleophilic, is favored. At pH 7.0 and 0 °C, for instance, hydrolysis half-life is 4 to 5 h. For this reason, protein labeling must needs employ a great excess of NHS ester.[1][2][4][5]
For applications routinely requiring derivatization of primary amines, NHS esters are advantageous for a number of reasons. Anderson et al.[2] describe that they are highly reactive and thus give good yields in amine couplings; moreover, being crystalline, they are easy to handle. Good yields persist even when conducting the coupling reaction in the presence of water, which sets NHS esters apart from several other active esters.[4] An important benefit of NHS esters is that, in their crystalline form, they are relatively resistant to hydrolysis and thus the concomitant inactivation, meaning that they can be stored for an extended period of time under dry conditions without special precautions while retaining their activity.[3] However, this comes with a caveat: solutions of NHS esters are susceptible to hydrolysis especially if prepared in N,N-dimethylformamide or dimethyl sulfoxide. These hygroscopic solvents tend to draw moisture, promoting hydrolysis of the NHS ester; such solutions should thus be prepared immediately before use.[6]
In our work, NHS esters containing a biotin moiety, biotin-NHS among them, are used to biotinylate inanimate protein components of bioink or reservoir, for instance PAS-Lysine. For the printed mesh to be stable, this covalently attached biotin must not be cleaved. In this respect, however, we now encounter a challenge: our printing system involves live cells. These cells require cell culture media, which commonly contain serum – and serum contains the potential downfall to mesh stability: the enzyme biotinidase.
How Stable is Biotin Coupled to Proteins?
Hydrolytic Cleavage
In the human body, biotin is a vital cofactor for several carboxylases, where it is attached to the ε-amino group of a lysine residue within the conserved sequence AMKM.[7][8] To liberate this covalently attached biotin, biotinidase, or biotin-amide amidohydrolase (EC 3.5.1.12), naturally hydrolyzes biocytin (ε-N-biotinyllysine) to biotin (vitamin H) and lysine.[7][9] The monomeric enzyme of size 76 kDa possesses a catalytic sulfhydryl group.[7][10] It not only serves to recycle biotin for renewed usage as well as to make protein-bound biotin in food bioavailable,[9] but also acts as a shuttle for biotin within the body[11]. Biotinidase deficiency thus causes multiple carboxylase deficiency, which, for one, entails serious developmental retardation because the body lacks the free biotin necessary to replenish its holocarboxylase pool; effective treatment is easily accomplished by administering daily doses of free biotin.[8]
Biotinidase is ubiquitous throughout the human body. Its specific activity is highest in serum (5.80±0.89 nmol min–1 mL–1[12]), followed by other organs including the liver, where the enzyme is produced.[8]
While biotinidase used to be considered fairly specific for biocytin,[9] evidence has accumulated that this is not the case[13]. Thus, it can also hydrolyze other biotin amide or ester conjugates, such as the methyl ester[14] or the conjugate with p-aminobenzoate, which forms the basis of a colorimetric activity assay[9][10].
Biotinidase is thought to primarily act on biocytin or small biotin-bearing oligopeptides in vivo, since its activity rapidly diminishes with increasing peptide length; it is ultimately unable to cleave biotin off of holocarboxylases.[7] However, other studies have shown that the enzyme is capable of acting on intact proteins, such as biotinylated IgG[15] and histones[16]. Since biotin participates in the chemical reaction catalyzed by carboxylases and is thus somewhat shielded, this discrepancy likely stems from different exposure to the surrounding medium.
The issue is this: With all biotin attached to lysine on both cell receptors and spacer molecules, we will invariably face slow hydrolysis if a cell mesh is exposed to serum-containing media or applied in vivo. Depending on the task at hand, we might desire our mesh to either be stable long-term or to be disintegrated more quickly. The bonding mode of the spacer’s biotin, which is introduced chemically, is key for driving this in either direction.
Stabilizing the Biotin Peptide Bond
If we are to prevent biotinidase from cleaving biotin off of biotinylated components in our bioprinting system and thus from disintegrating our mesh while simultaneously obviating use of special serum-free media, we must consider how to render the biotin bound to the spacer peptide non-hydrolyzable by biotinidase.
Biotinidase-resistant biotin conjugates have been extensively studied in the context of the cancer treatment "pretargeting" approach. Here, biotinylated IgG is bound to the tumor followed by sequential treatment with streptavidin and radionuclide-containing biotin, which offers the advantages of fast accretion as well as clearance combined with low tissue retention (Bogusiewicz et al., 2004; Goldenberg, Chatal, Barbet, Boerman, & Sharkey, 2007).[15][17] The general consensus is that steric hindrance at and adjacent to the peptide bond to be cleaved is paramount; for instance, p-aminobenzoate is cleaved off much faster than the m-isomer, and the o-isomer is not cleaved at all. Thus, resistance is acquired either if the amide nitrogen is tertiary, e.g. carrying an additional methyl group (as in biotinyl-sarcosine), or if the position next to this nitrogen carries a substituent.[18] Among the reported non-hydrolyzable biotin derivatives are those with the following substituents adjacent to the amide nitrogen: hydroxymethyl (serine), carboxylate (aspartate), ethyl (2-aminobutyric acid), isopropyl (valine), benzyl (phenylalanine); a methyl group (alanine), however, is not sufficient.[13][9]
Since increased steric bulk diminishes the affinity of streptavidin toward the biotin, the 2-aminobutyric acid derivative has been preferred over e.g. the valine conjugate because it binds to streptavidin more strongly and is less lipophilic. However, this deviation may not ultimately be relevant given the inherent exceptionally high avidity of streptavidin toward biotin.[13]
Design
Experiments
Proof of concept
Demonstrate
Discussion
References
- ↑ 1.0 1.1 1.2 Hermanson, G. T. (2013) Bioconjugate Techniques (3 ed.). Boston: Academic Press.
- ↑ 2.0 2.1 2.2 2.3 Anderson, G. W., Zimmerman, J. E., & Callahan, F. M. (1963). N-Hydroxysuccinimide Esters in Peptide Synthesis. Journal of the American Chemical Society, 85(19), 3039-3039.
- ↑ 3.0 3.1 3.2 Klykov, O., & Weller, M. G. (2015). Quantification of N-hydroxysuccinimide and N-hydroxysulfosuccinimide by hydrophilic interaction chromatography (HILIC). Analytical Methods, 7(15), 6443-6448.
- ↑ 4.0 4.1 Anderson, G. W., Zimmerman, J. E., & Callahan, F. M. (1964). The Use of Esters of N-Hydroxysuccinimide in Peptide Synthesis. Journal of the American Chemical Society, 86(9), 1839-1842.
- ↑ Kalkhof, S., & Sinz, A. (2008). Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Analytical and Bioanalytical Chemistry, 392(1), 305-312.
- ↑ Savage, M. D. (1996). An Introduction to Avidin-Biotin Technology and Options for Biotinylation. In T. Meier & F. Fahrenholz (Eds.), A Laboratory Guide to Biotin Labeling in Biomolecule Analysis. Basel: Birkhäuser.
- ↑ 7.0 7.1 7.2 7.3 Craft, D. V., Goss, N. H., Chandramouli, N., & Wood, H. G. (1985). Purification of biotinidase from human plasma and its activity on biotinyl peptides. Biochemistry, 24(10), 2471-2476.
- ↑ 8.0 8.1 8.2 Wolf, B. (2010). Clinical issues and frequent questions about biotinidase deficiency. Molecular Genetics and Metabolism, 100(1), 6-13.
- ↑ 9.0 9.1 9.2 9.3 9.4 Wolf, B., Hymes, J., & Heard, G. S. (1990). [11] Biotinidase. In W. Meir & A. B. Edward (Eds.), Methods in Enzymology (Vol. 184, pp. 103-111): Academic Press.
- ↑ 10.0 10.1 Knappe, J., Bruemer, W., & Biederbick, K. (1963). Reinigung und Eigenschaften der Biotinidase aus Schweinenieren und Lactobacillus casei. Biochemische Zeitschrift, 338, 599-613.
- ↑ Chauhan, J., & Dakshinamurti, K. (1988). Role of human serum biotinidase as biotin-binding protein. Biochemical Journal, 256(1), 265-270.
- ↑ Wolf, B., Grier, R. E., Secor McVoy, J. R., & Heard, G. S. (1985). Biotinidase deficiency: A novel vitamin recycling defect. Journal of Inherited Metabolic Disease, 8(1), 53-58.
- ↑ 13.0 13.1 13.2 Wilbur, D. S., Hamlin, D. K., & Chyan, M.-K. (2006). Biotin Reagents for Antibody Pretargeting. 7. Investigation of Chemically Inert Biotinidase Blocking Functionalities for Synthetic Utility. Bioconjugate Chemistry, 17(6), 1514-1522.
- ↑ Pispa, J. (1965). Animal Biotinidase. Annales Medicinae Experimentalis et Biologiae Fenniae, 43, 1-39.
- ↑ 15.0 15.1 Bogusiewicz, A., Mock, N. I., & Mock, D. M. (2004). Release of biotin from biotinylated proteins occurs enzymatically and nonenzymatically in human plasma. Analytical Biochemistry, 331(2), 260-266.
- ↑ Ballard, D. T., Wolff, J., Griffin, B. J., Stanley, S. J., van Calcar, S., & Zempleni, J. (2002). Biotinidase catalyzes debiotinylation of histones. European Journal of Nutrition, 41(2), 78-84.
- ↑ Goldenberg, D. M., Chatal, J.-F., Barbet, J., Boerman, O., & Sharkey, R. M. (2007). Cancer Imaging and Therapy with Bispecific Antibody Pretargeting. Update on cancer therapeutics, 2(1), 19-31.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedchauhan1986