Contents
- 1 Attribution
- 2 Finding the biotINK project
- 3 Our principal investigators (PIs)
- 4 Our Journey Through Munich
- 5 Attributions sorted by methods
- 5.1 Protein Production and Purification
- 5.2 Molecular Cloning
- 5.3 Western Blot
- 5.4 Mass spectrometric measurements
- 5.5 Determination of mRNA levels with RT-q-PCR
- 5.6 Incubation facility
- 5.7 Flow Cytometry
- 5.8 Fluorescence Microscopy
- 5.9 Scanning Electron Microscopy
- 5.10 Blood samples
- 5.11 Chemical synthesis of Biotin-NHS active esters
- 5.12 Maya Video
- 5.13 Micromanipulator experiments
- 5.14 Business plan
- 5.15 Survey: "Supporting Entrepreneurship"
- 5.16 The Wiki
- 5.17 Video cutting
Attribution
Attributions are an important point of any iGEM project as it is necessary to put the results obtained by the team into the context of individual responsibilities. Thus, we will attempt to candidly describe how our project evolved, which student was responsible for which sub-project, and who supported our project with scientific or non-scientific contributions.
Finding the biotINK project
For the story of finding our project, it is essential to know that there was no iGEM team from either TUM or LMU in 2015 and no team at TUM in 2014. Thus, each of these years saw students wanting to participate in the iGEM competition but facing the bleak perspective that there was no hosting lab that offered the support necessary to make the "iGEM experience". For this long a time, students have brainstormed in order to find interesting ideas that they could convert into an iGEM project. Likewise, the idea for our bioprinting project is far older than just this year – at least in theory. The life science students finally assembled around last February, eagerly suggesting various ideas for several promising potential iGEM projects that they had in mind. The project ideas that were developed most involved i) transforming the planet Mars with bacteria, ii) creating a vegan substitution product for milk, and iii) bioprinting. While we considered the bioprinting project to be the most intriguing idea, last year's TU Delft iGEM team had unfortunately already developed an excellent project on bioprinting. Thus began a long discussion of whether it would be in our team's best interests to participate with another bioprinting project. In the end, however, we agreed that the next years of iGEM will most likely witness the establishment of a separate bioprinting track and that this topic has extraordinary potential. By the way, it was much encouraging to listen to Randy in Paris when he described the potential of SynBio and chose bioprinting of organs for an example! At this point, our last doubts melted, and we were wholly confident that our project choice was just right.
Our principal investigators (PIs)
As implied above, it was very difficult to convince a professor at TUM of an iGEM participation, and at LMU as well it was cumbersome to find a PI enthusiastic about iGEM now that Prof. Mascher has left Munich. The initial motivation to found this year's iGEM team was sparked by the Graduate Center GRK 2062: Molecular principles of synthetic biology, which desired to re-establish the iGEM competition in Munich.
- Prof. Dr. Jung (one of our secondary PIs, LMU) is the head of this graduate center and thus impulsed the creation of a team by recruiting students. Wanting to share the project she (and the GRK) looked for a lab participating in the graduate center that could host the iGEM team as main lab. During the summer we had access to lab space in her department, where we could perform cloning steps and other work. This was of great help, as the distance between LMU and TUM at Weihenstephan Campus is more than 2 h with public transportation. We are grateful for Prof. Jung's support even though towards the end we decided to rejoin into our main lab.
- Prof. Dr. Skerra (our main PI, TUM) already filled the role of the main PI of TUM's 2012 and 2013 iGEM teams and thus had the first-hand experience of what it means to host an iGEM team. Initially, he was disinclined to host yet another iGEM team at his lab, but finally we could convince him of this. He is a renowned expert in the field of protein design, especially for therapeutic proteins, and has also founded two successful companies. For this reason, he is quite busy and unable invest great amounts of time in the iGEM competition. He joined our weekly team meeting once after we had decided on our project, giving us several invaluable tips concerning what components to integrate and what aspects to focus on in order to arrive at a proof of principle in the end. Apart from this session with him, it is his philosophy that iGEM is a student's competition and that the students themselves should design and create the project. He generously offered all of the resources available at his chair together with the excellent analytical equipment, while letting us free rein to plan and execute the project on our own – and we are very grateful that he provided us with this opportunity!
- Prof. Dr. Westmeyer (secondary PI, TUM, Helmholtz center) was asked to join the team halfway through the project when we realized that we required his expertise in the field of fluorescence microscopic imaging. He supported us, especially in the final weeks, with cell-biological reagents for staining and transfection as well as with the analytical instrumentation of his lab. As he spent the last years in Boston at the MIT and at Harvard, he is very open-minded concerning the potential and possible applications of SynBio in the future.
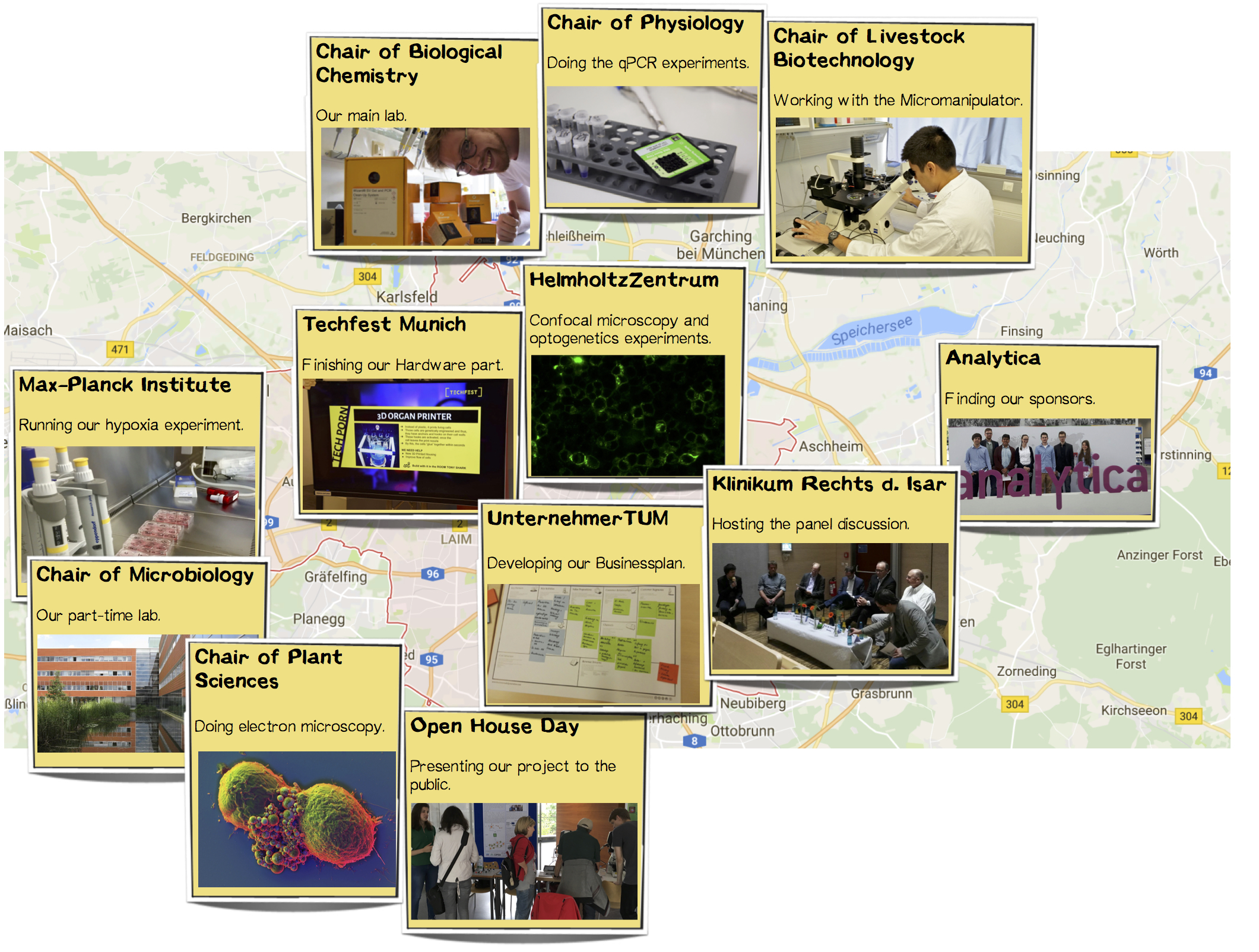
Our Journey Through Munich
In the course of our project, we have met many nice people willing to help and support us with lab space, knowledge, or materials. Below you can find all the stations we stopped by throughout our journey up to the outcome of our project.
Attributions sorted by methods
All work associated with planning, conducting, and evaluating experiments was performed by student members of our team. Instructors assisted the students in planning their experiments, introducing relevant molecular-biological and chemical techniques to the students when they performed a specific experiment for the first time. Moreover, the students evaluated the experiments themselves after an introduction to the theory and advice on what to do and what to avoid. After this preliminary evaluation, all data were accumulated and discussed in the weekly team meeting, at which our two instructors Andreas and Volker (both PhD students at the lab of Prof. Skerra) were present to reflect on our project. Here we enumerate the specific tasks as well as the student(s) who performed them in order to allow the judges to directly contact the student responsible for a topic of interest.
Protein Production and Purification
As we produced, purified, conjugated, and analyzed many different protein samples, several team members were involved, including Niklas, Jan and Julian. Shaker flask productions were conducted independently of other lab members; for fermentation (which turned out to be a somewhat more difficult than explained in lectures), one of our instuctors had an eye on the whole production. Purification with normal columns was performed independently; for those involving the Äkta purifier, a lab member supervised our handling for the first 3 to 5 chromatographies.
Molecular Cloning
In the first week, we received an introduction to molecular cloning. Afterwards, we worked independently of our instructors and only asked them from time to time whenever a problem occurred. The gene design for all BioBricks was conceptualized by a student member (often Clemens or Christoph) and then reviewed by our instructor Volker.
Western Blot
All Western Blots were done by Luisa, and she received an introduction to blotting and staining from Fabian Rodewald and Christian Deuschle.
Mass spectrometric measurements
The correctness of recombinant proteins before and after conjugation was analyzed by ESI-TOF mass spectrometry. Samples were prepared by student members of the team, the actual measurement was performed by a technical assistant (which is also true for PhD students).
Determination of mRNA levels with RT-q-PCR
To further characterize the expression of the different receptor constructs our team wanted to take a closer look at the respective mRNA levels. In order to do so we asked Professor Dr. Pfaffl for help, since he is one of the leading experts for quantitative PCR. Together with him we were able to find the right approach to solving our problem. We want to thank Professor Dr. Pfaffl for his advice and for supplying the lab space for conducting our experiments. We also want to thank one of his PhD students, Dominik Buschmann, for helping us with the experiments and the analysis of the gathered data.
Incubation facility
The hypoxia experiments could only be performed thanks to the support of the Schwille Group at the Max-Planck Institute of Biochemistry in Martinsried, which kindly provided the required incubation facility. In this context, we are indebted to Tobias Härtel and Bea, who always offered their invaluable advice and help. The hypoxia experiments were performed by Jan and Elena, who were ready to take samples every 12 hours.
Flow Cytometry
The flow cytometry experiments were all performed by Luisa as she spent 18 months in a research group in Hamburg before she started her studies in Munich, thus already versed in all skills necessary to measure our cells in FACS.
Fluorescence Microscopy
Fluorescence microscopy at the lab of Prof. Skerra was performed by different people as it was necessary for many different sub-projects. High-resolution confocal fluorescence microscopy was conducted at the Helmholtz Center in Munich in the lab of Prof. Westmeyer by Christoph as he will also do his master's thesis in this lab. He received the introduction into staining cells and performing confocal imaging experiments from our external Instructor Jeff.
Scanning Electron Microscopy
Our colorful Scanning Electron Microscopy images were shot by Prof. Dr. Wanner from the Plant Science Department of the Ludwig-Maximilians University. Elena approached him to support our project in terms of a collaboration to provide us impressive electron microscopy images of cells. Prof. Wanner supplied all reagents required for sample preparation, such as microscopic slides, glutaraldehyde, transport boxes and many more! Clemens was in charge of preparing the samples late in the evening, which were thus slightly improvised. The samples were then picked up and transported by Jan and Elena to the SEM facility. Afterwards, the remaining sample preparation steps after the fixation were carried out by Prof. Wanner and his coworkers. We are sincerely grateful for his support!
Blood samples
For our "linker chemistry" subproject, we needed fresh blood samples as a readily available source of human biotinidase in the form of plasma. For this purpose, Vivien, the student who performed the chemical experiments, utilized her own blood graciously drawn by PD Dr. med. Thomas Skurk, a medical doctor at the Institute for Food & Health.
Chemical synthesis of Biotin-NHS active esters
Chemical synthesis was performed by Vivien, as she wrote her bachelor's thesis at the Chair of Organic Chemistry at TUM. The initial synthesis plan was discussed with Dr. Martin Dauner, who is the chemist in the lab of Prof. Skerra. Vivien worked alone afterwards in synthesizing the target compounds and subsequently analyzing their properties. The NMR experiments were then measured at the Faculty of Chemistry in Garching. Vivien evaluated all results (including NMR).
Maya Video
To be honest, we are very proud that the Maya Video Vivien built looks so great in the end. Initial renderings that she showed to the team were far more simple than the final product. She had already gathered some experience in 3D animating, having regularly worked with Blender as a hobby before; this helped her very much in understanding the Maya software faster (which is really not that intuitive). She also compiled a How-To Video in which she explains the creation of the video in just short of one hour. If you are interested in the video, please ask her, as she really did it on her own ;)
As there was no PDB file available from the PAS-polypeptide we asked the Junior-PI Dr. Andre Schiefner at the Chair of Biological Chemistry (TUM) for help. As an experienced christallographer he was able to create a PAS(200) sequence with lysines that could be used later on for the creation of the PAS animation.
Micromanipulator experiments
We thank Professor Dr. Schnieke and her [http://btn.wzw.tum.de/Chair of Livestock Biotechnology] at the WZW (TUM School of Life Sciences Weihenstephan) for so generously providing us with the opportunity to use the micromanipulator at the chair's laboratory and allowing us to conduct several highly specific experiments with our bioink. With the extensive help and instructions of Dr. Nicolas Ortega, a researcher at the lab having much experience with the device, we were able to plan experiments and test our bioprinting technique on a minuscule scale. In the process, our team members Julian and Javier were able to observe the ejection process under controlled conditions in great detail and document the behavior of our modified HEK cells containing ink. Thanks to this opportunity we could obtain useful and valuable visual material as well as a better understanding of our biotINK concept.
Business plan
The business plan was written by Christoph as he also studies TUM Economics besides his studies in Molecular Biotechnology.
Survey: "Supporting Entrepreneurship"
For the creation of this survey, we had a special Instructor, Philipp. He is a PhD student an the Chair of Technology and Innovations Management. He supervised Hallie, an exchange student from the USA who joined our team to create this survey and elucidate how we can make iGEM more productive concerning its output in successful start-up companies.
The Wiki
The Wiki was programmed by Peter, a student in Media Informatics (LMU). As we encountered some problems finding someone to program the Wiki, he joined the team 4 weeks before wiki freeze. As he also works in web design and completed his bachelor's thesis in parallel, he was quite busy and did not have overly much time (which is why we finalized our CSS on the last evening). In the end, though, he successfully completed the work.
Video cutting
All work associated with recording as well as processing auto and video files was done by Julian.