(→Medical Application) |
(→Medical Application) |
||
| Line 3: | Line 3: | ||
=Medical Application= | =Medical Application= | ||
| − | ==Therapeutic Application ≠ usage of fancy state-of-the-art methods | + | ==Therapeutic Application ≠ usage of fancy state-of-the-art methods but reliable simplistic methods== |
<div class="white-box"> | <div class="white-box"> | ||
| − | In | + | In biomedical Research, only transparent and simplistic therapeutic approaches, which can be evaluated in all stages of clinical trials, will end up in the patient. |
Nowadays, one of the holy grails in Synthetic Biology is a) to understand the complex regulation mechanism on (co-)transcriptional and (co-)translational networks on every imaginable level and b) to manipulate and rewire those networks. | Nowadays, one of the holy grails in Synthetic Biology is a) to understand the complex regulation mechanism on (co-)transcriptional and (co-)translational networks on every imaginable level and b) to manipulate and rewire those networks. | ||
| Line 13: | Line 13: | ||
Great efforts, especially in Computational Biology and Systems Biology, have been achieved to understand the complex homeostasis-network of life. Although many principles of regulation have been understood, only relative small networks can be modelled and simulated computationally; the outcome of a “healthy” network are sometimes well understood in this case. | Great efforts, especially in Computational Biology and Systems Biology, have been achieved to understand the complex homeostasis-network of life. Although many principles of regulation have been understood, only relative small networks can be modelled and simulated computationally; the outcome of a “healthy” network are sometimes well understood in this case. | ||
| − | + | But pathological networks, where a defective network node causes a shift in the cell/organ/organisms’ homeostasis and thus result in “disease” are poorly understood, since there are still too many unknown network nodes and/or the dynamic and impact of a node is not understood. | |
Consequently, the rewiring of a complex pathological network to achieve a desired outcome is even more complex due to many unknown variables; thus, it is most likely illusional to only rely on rewired genetic circuits on engineered cells to achieve healing in an individual. | Consequently, the rewiring of a complex pathological network to achieve a desired outcome is even more complex due to many unknown variables; thus, it is most likely illusional to only rely on rewired genetic circuits on engineered cells to achieve healing in an individual. | ||
Revision as of 01:49, 20 October 2016
Medical Application
Therapeutic Application ≠ usage of fancy state-of-the-art methods but reliable simplistic methods
In biomedical Research, only transparent and simplistic therapeutic approaches, which can be evaluated in all stages of clinical trials, will end up in the patient.
Nowadays, one of the holy grails in Synthetic Biology is a) to understand the complex regulation mechanism on (co-)transcriptional and (co-)translational networks on every imaginable level and b) to manipulate and rewire those networks.
Great efforts, especially in Computational Biology and Systems Biology, have been achieved to understand the complex homeostasis-network of life. Although many principles of regulation have been understood, only relative small networks can be modelled and simulated computationally; the outcome of a “healthy” network are sometimes well understood in this case.
But pathological networks, where a defective network node causes a shift in the cell/organ/organisms’ homeostasis and thus result in “disease” are poorly understood, since there are still too many unknown network nodes and/or the dynamic and impact of a node is not understood.
Consequently, the rewiring of a complex pathological network to achieve a desired outcome is even more complex due to many unknown variables; thus, it is most likely illusional to only rely on rewired genetic circuits on engineered cells to achieve healing in an individual.
Of course, it will just be a matter of time until engineered cells can sense the “problem”, to then integrate and compute it accordingly generating an output function to “solve the problem”. Unfortunately, for now this still remains science fiction.
Biologization of Technology vs. Technologization of Biology
Currently, there is a trend to solve biological and even technical problems with Biology (Biologization of Technology), especially Synthetic Biology, e.g. engineered algae fuel or solve mathematical problem (traveling salesman problem as popular example) with swarming bacteria (Myxococus) or Mycetozoa; unquestionably, those great results are of major impact to solve or understand global challenges.
But in biomedical research, it is undisputed that many challenges in healthcare cannot just be solved with biology and needs interdisciplinary help from many scientific fields for reasons, mentioned above. Many (pathological) biological networks are too poorly understood and the construction of a “rewired correction network” to compensate the misregulated homeostasis is consequently unpredictable and thus not available for human therapy. Of course there a exceptions, where fully rewired cellular systems are currently in trial for humans, e.g. usiig Chimeric Antigen Receptors (CAR) on T-cells as cancer therapeutic.
Technical solutions to human health problem are more widely available (Technologization of Biology) since this discipline exists just longer thanSynthetic Biology. The most-popular examples are fully non-biological solutions, e.g. an artificial heart, which can replaces the human heart for a long time. The reasons why this solution can be be “just technical” is that the human heart is basically just a very complicated pump. The challenges were more of fluiddynamic nature to pump our blood without destroying or clotting the cells.
But there a many medical problems which cannot be solved by just using parts of electromechanical engineering. A showcase would be glucose-resistant islet cell (one of the many forms of diabetes) which does not secrete insulin when stimulated with glucose. No non-biological solution will ever exist for this, since the only input function to bring back the dysregulated metabolism back to the physiological homeostasis is the protein insulin, an undoubtedly biological component.
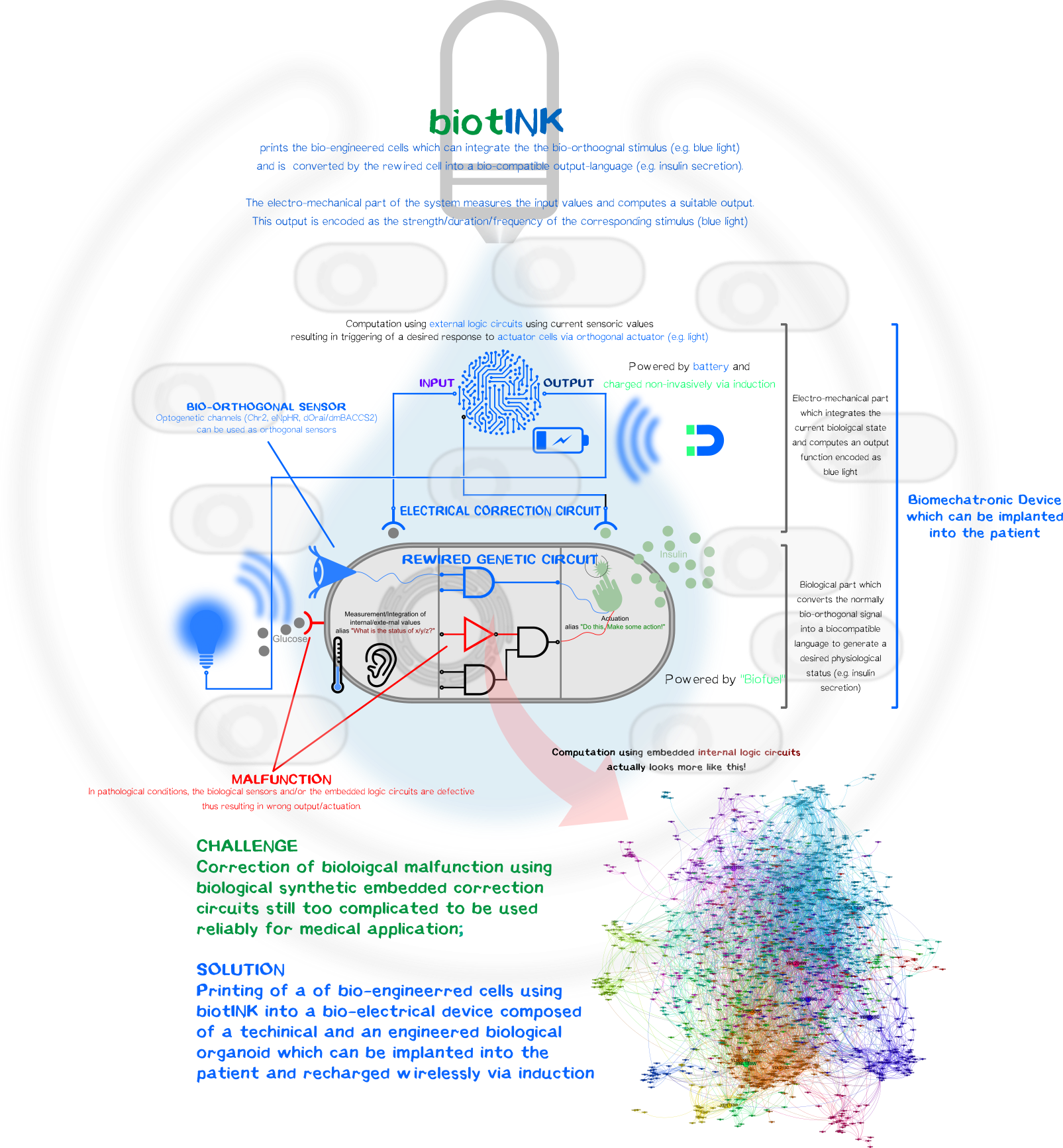
A kybernetic bio-printed device to control blood glucose level
However, one can luckily combine the best of both disciplines, the reliablity and durability of electromechanical components and the ability to create simplistic bio-orthogonal signal transducer/integrator to capture the output function and translate it into a bio-compatible signal.
To remain with the above mentioned example with glucose-resistant or destroyed β-cells by autoimmune disease, one in general could bio-print (LINKLINKLINKLINKLINKLINKLINKLINKLINKLINKLINKLINKLINK click here for more on our bioprinting project) an kybernetic organ composed of a electromechanical part and a biological part. The technical part would measure the current physiological state of the body (glucose level) and will compute an output function encoded as a normally bio-orthogonal signal; only the desired printed target cells will respond to this signal (Bio-orthogonality minimizes the off-target effects compared to the usage non-bio-orthogonal signals). This signal will be integrated intracelluarly resulting in the generation of a desired biological output function to bring back or maintain the glucose homeostasis. The change is again measured by the electronic sensor which then again generates a new output function for the bio-engineered cells. The electromechanical, bio-engineered and biological parts form together a so-called closed-loop-control-circuit.
Biological Part
Optogenetic Control of Calcium-Signaling
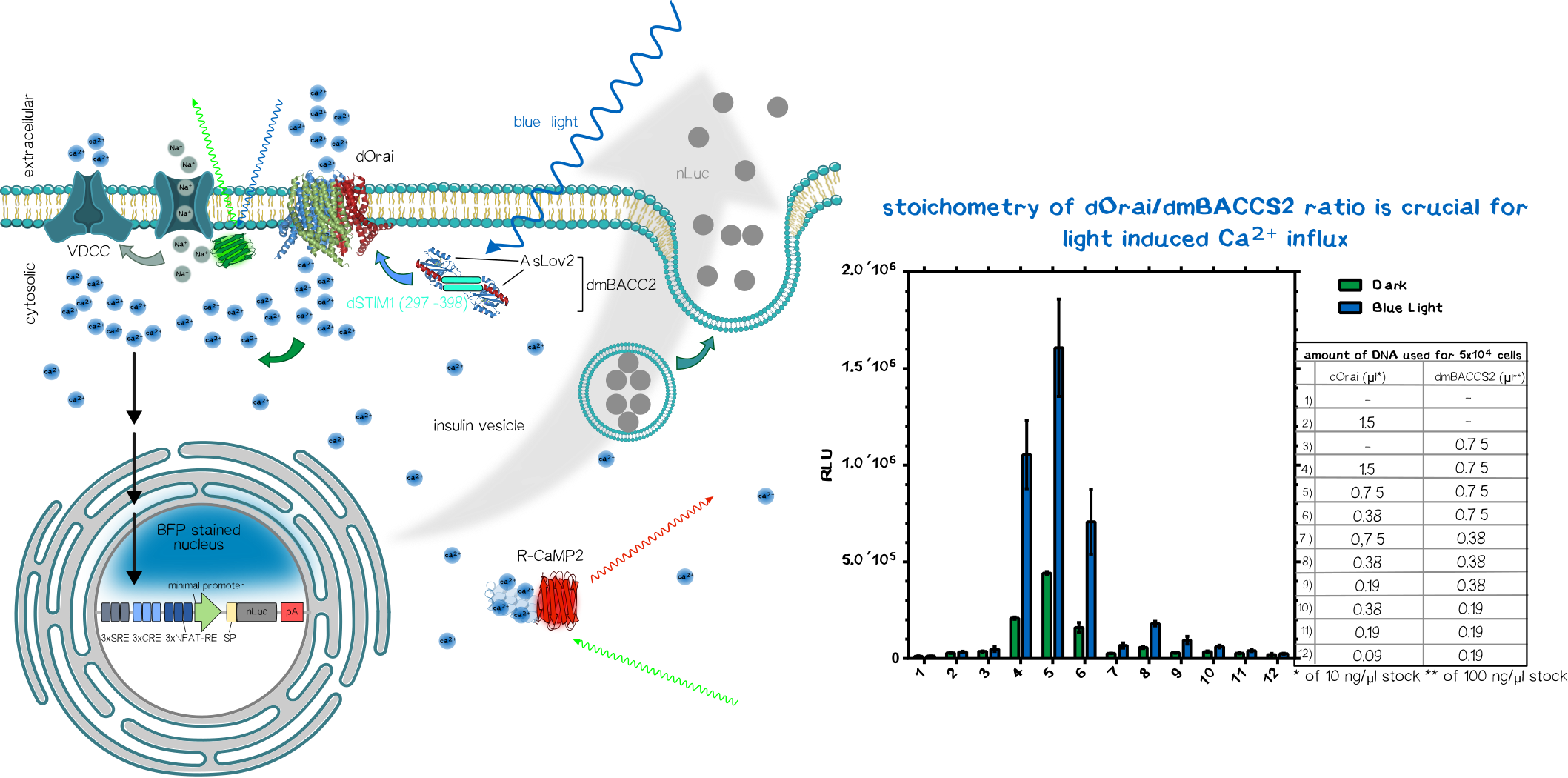
To sense the normally bio-orthogonal output blue light (~450 nm) (of course there are photoreceptors in our retina which can sense blue light but normal visceral cells normally does not react to blue light), one have to bring in a sensoric system which converts the electro-magnetic signal in the visible range into a bio-compatible signal. The desired second messenger here is Ca2+.
We made usage or the recently published system (REFERENCEREFERENCEREFERENCEREFERENCEREFERENCEREFERENCE) using the calcium-selective channel Orai1 and the Orai1 interaction motif from Stim1. Stim1 is a ER-resident single-pass transmembrane protein which has a lumenal calcium sensing part and a cytosolic part that can activates Orai1 and thus mediating the influx of of Ca2+ ions. This mechanism is triggered when the ER calcium concentration is low, resulting the conformational change of the lumenal EF1-hand motif of Stim1. This change induces the clusterting of Stim1 resulting in the relocation of Stim1 near the plasma membrane (PM). There it interact with the PM-located Orai1 and induces Orai1-mediated Ca2+ influx. The cytosolic calcium ions are then used to refill the ER-Ca2+-storage.
In the system we used, Orai and Stim1 is derived from Drosophila melanogaster for additive bio-orthogonality (mammalian and Drosophila Orai1/Stim1 system are not interchangeable). The Orai1 of D. melanogaster is used in its wildtype form (dOrai). The D. melanogaster is fused to the C-terminus of the an engineered variant the the phototropin LOV2-Jα domain from Avena sativa (Chimeric protein is called dmBACCS2). Under basal condition (without light), the Stim1 peptide is quenched by the Jα-helix; however, under illumination with blue light, the Jα-helix unwinds and the Stim1 peptides can multimerize and mediates Ca2+-influx via dOrai1.
Thus, the dOrai/dmBACCS2 system can be understood as a blue-light-to-Ca2+-converter. Afterwards we have to translate this bio-compatible Ca2+-signal into a biological output function.
Integration of Calcium-Signaling
The output function of the technical parts encoded as the modulation of frequency/amplitude/pulse duration of the blue light is already converted into a biocompatible Ca2+-signal which now have to integrated into an output function.
We made usage of a simplistic system by putting multiple transcription factor site, known for their nuclear translocation and binding to their DNA target motifs upon calcium singaling, upstream of a minimal promoter sequence. Those motifs were Serum Response Elements (SRE), cAMP Response Elements (SRE) and NFAT Response Elements (NFAT-RE) which are well-known of the calcium-mediated effects. For the aforementioned case of Diabetes, one would put the coding region of preproinsulin.
As proof-of-concept we put an secreted nano-luciferase downstream after the minimal promoter instead for reasons of easy quantification. Thus the luminescence in the collected supernatant allows us to quantify how much protein is secreted within a defined time-frame.
Results
After co-transfection of the dOrai/dmBACCS2 system together with a secreted nanoluciferase reporter containing response elements of calcium signaling preceding a minimal promoter, one can observe a clear increase in signal in the illuminated samples compared to the non-illuminated ones. After 24 of blue light (~450 ), the amount of supernatant nanoluciferase is about 4-5 times higher compared to the non-illuminated control condition.
In parallel we also tested the meGFP-tagged channelrhodopsin-2 (Chr-2 H134R mutant) to activate the voltage-dependent calcium channels (VDCC) to generate the same effect. The genetically encoded Ca2+ indicator was also on the same plasmid including a nuclear-localized blue fluorescent protein mTagBFP2. After transfection we could see nicely the Chr2-H134R localization on the PM and also the nuclear localized mTagBFP2 (Figure 3). When exposed to blue light, we could not observe an increase in red fluorescence due to the limited temporal resolution of our eppifluorescence microscope. Moreover, we could not see an increase in nanoluciferase signal after 24 of illumination in contrast to the above mentioned dOrai/dmBACCS2 system (data not shown).
Technical Part
Bio-Printing
LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE LINK ZU BIOPRINT SEITE
Wireless Charching
VERWEIS AUF EXISTIERENDE LÖSUNGEN WIE QI LADEGERÄTE FÜR HANDYS ETC.
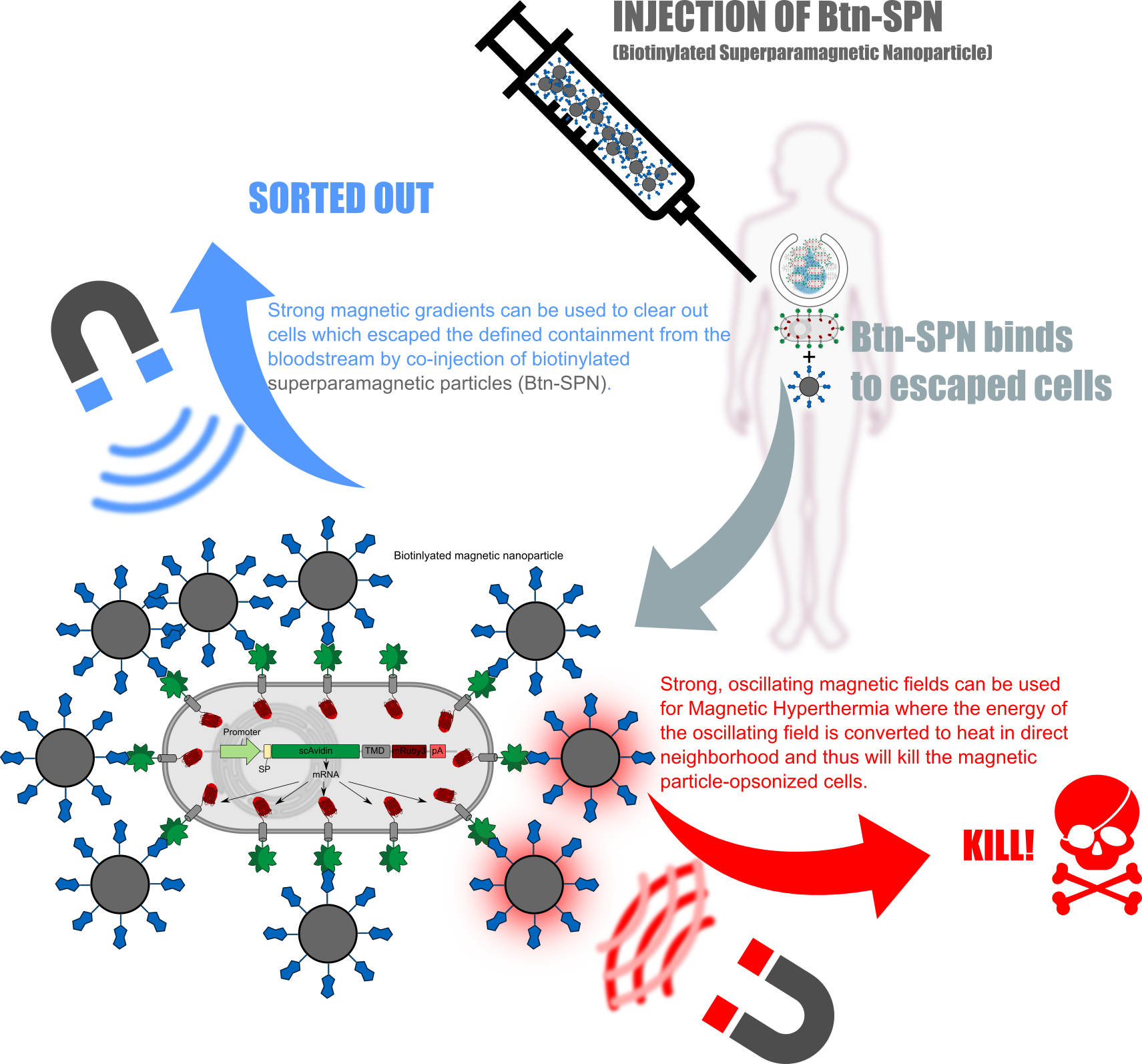
Kill two birds with one stone
QUERVERWEIS AUF ALLE ANDEREN UNTERSEITEN; WIE POLYMERISATION; RECEZPTOR; BIOPRINT ETC:
ERKLÄREN WARUM REZETPRO GEILER SCHEISS IST; DA EINERSEITS STRUKTUR GIBT UND ANDERERSEITS; FALLS DIESE AUSVERSEHEN IN DIE BLUTBAHN GELANGT MAN DURCH CONINEKTION MIT MAGNETIC PARTICLES SCHNELL WIEDER RAUSFISCHEN KANN ODER DIREKT TÖTEN KANN!
Literaturreferenz
Literaturreferenz[1]
Bei Google Scholar bitte das APA-Ziteirformat verwenden.
Textformatierung
kursiv
fett
Strich
Links
Wikiinterner Link Team:LMU-TUM_Munich/Materials (As described in the Materials section)
Wikiexterner Link Visit W3Schools
Visit W3Schools
Bilder
Introduction
Design
Experiments
Proof of concept
Demonstrate
Discussion
References
- ↑ Schmidt, T. G., & Skerra, A. (2007). The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nature protocols, 2(6), 1528-1535.