Javier.luna (Talk | contribs) (→Results: Polymerization experiments) |
Javier.luna (Talk | contribs) |
||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
[[File:Muc16_Sticker_Proof_001.png |right|350px]] | [[File:Muc16_Sticker_Proof_001.png |right|350px]] | ||
| − | + | =Design: Why we chose the Avidin & Biotin affinity pair= | |
| − | + | We have discussed a long time what kind of biochemical interaction we could use to have a rapid and high-affinity interaction of the components. Factors that influenced our choice were: affinity (K<sub>D</sub>), association rate (k<sub>on</sub>) as well as the number of binding sites we can have on a recombinant protein as well as the yield it gives when produced recombinantly. The comparison with some other possible affinity-pairs is shown in the following table.<br> | |
| − | |||
| − | |||
| − | |||
| − | |||
{| | {| | ||
|+ Properties of membrane proteins | |+ Properties of membrane proteins | ||
| Line 34: | Line 30: | ||
=Multivalency and avidity as major principles for polymerization= | =Multivalency and avidity as major principles for polymerization= | ||
| − | + | A nice point about the biotin - Avidin affinity pair is that biotin groups can be conjugated to any protein of choice that contains surface-exposed aminogroups. In the next line we have visualized aminogroups in two proteins that we have used in our project as they are available in big amounts: enhanced Green Fluorescent Protein (eGFP, left) and Bovine Serum Albumine (BSA, right)[[File:Muc16-eGFP-annimatedGIF.gif]] | |
| − | [[File:Muc16-eGFP-annimatedGIF.gif]] | + | |
[[File:Muc16-BSA-annimatedGIF.gif]] | [[File:Muc16-BSA-annimatedGIF.gif]] | ||
| Line 42: | Line 37: | ||
Since the foundation of our polymerization reaction is the interaction of Streptavidin and its binding partner Biotin (Referenz) we used biotinylated polymers to convery the polymerization between the cells. | Since the foundation of our polymerization reaction is the interaction of Streptavidin and its binding partner Biotin (Referenz) we used biotinylated polymers to convery the polymerization between the cells. | ||
| − | [[File:MUC16_polymerization_001.png|thumb|center| | + | [[File:MUC16_polymerization_001.png|thumb|center|500px]] |
=Results: Polymerization experiments= | =Results: Polymerization experiments= | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Creating the perfect mesh== | ==Creating the perfect mesh== | ||
| Line 101: | Line 82: | ||
== Sticky cells in the Micromanipulator == | == Sticky cells in the Micromanipulator == | ||
| − | To get a clearer view on how the cells polymerize we wanted to see the actual '''polymerization event''' and how the cells stick to each other. In order to do so | + | To get a clearer view on how the cells polymerize we wanted to see the actual '''polymerization event''' and how the cells stick to each other. In order to do so we used a micromanipulator for ejection of single cells. We compared the ejection of our '''biotINK''' in Streptavidin solution with the ejection in phosphate buffered saline (negative controle) to make sure that, if polymerization occures, it is because of an specific interaction of the cells with biotinylated BSA and streptavidin. |
| − | It was very interesting to see that there really is a '''difference''' between the two experiments in a way that the biotINK, ejected in streptavidin, contrary to the one ejected in PBS, does show '''polymerization of | + | It was very interesting to see that there really is a '''difference''' between the two experiments in a way that the biotINK, ejected in streptavidin, contrary to the one ejected in PBS, does show '''polymerization of cells'''. |
<html><p><video width=49% style="float:left; margin-top : 15px"; margin-bottom: 15px"; controls preload:"auto"> | <html><p><video width=49% style="float:left; margin-top : 15px"; margin-bottom: 15px"; controls preload:"auto"> | ||
Revision as of 02:57, 20 October 2016
Design: Why we chose the Avidin & Biotin affinity pair
We have discussed a long time what kind of biochemical interaction we could use to have a rapid and high-affinity interaction of the components. Factors that influenced our choice were: affinity (KD), association rate (kon) as well as the number of binding sites we can have on a recombinant protein as well as the yield it gives when produced recombinantly. The comparison with some other possible affinity-pairs is shown in the following table.
| Affinity pair | Kind of interaction | Dissociation constant | Association rate |
| Avidin - Biotin | non covalent | 10-15M [1] | 55 440 |
| SpyCatcher - SpyTag | covalent | not applicable | far too slow for rapid polymerization |
| StrepTactin - Strep-tag II | non covalent | ~10-6M[2] | affinity is too low for a constant interaction |
Multivalency and avidity as major principles for polymerization
A nice point about the biotin - Avidin affinity pair is that biotin groups can be conjugated to any protein of choice that contains surface-exposed aminogroups. In the next line we have visualized aminogroups in two proteins that we have used in our project as they are available in big amounts: enhanced Green Fluorescent Protein (eGFP, left) and Bovine Serum Albumine (BSA, right)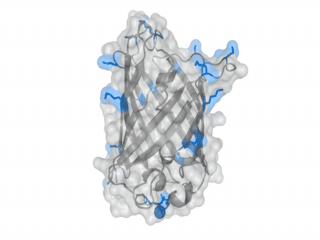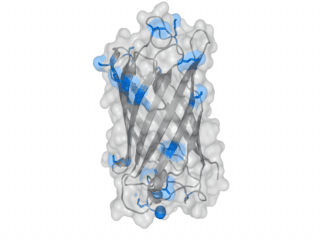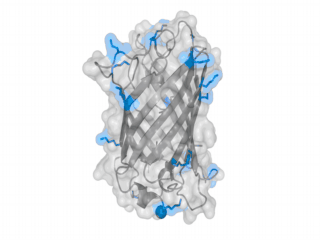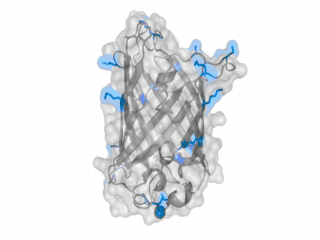

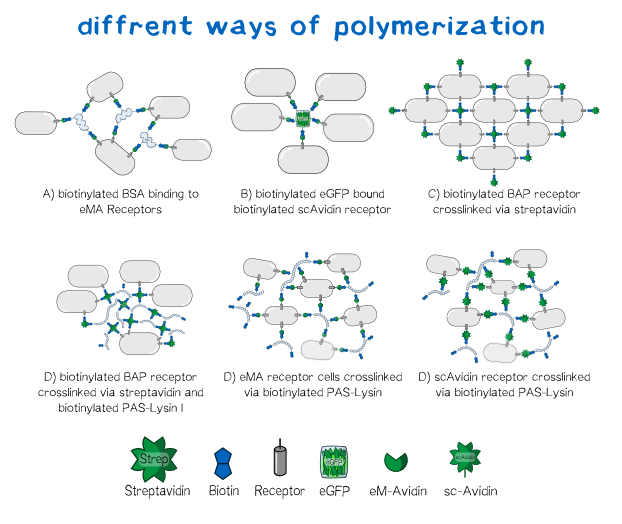
Different polymers possible with biotINK polymerization
Since the foundation of our polymerization reaction is the interaction of Streptavidin and its binding partner Biotin (Referenz) we used biotinylated polymers to convery the polymerization between the cells.
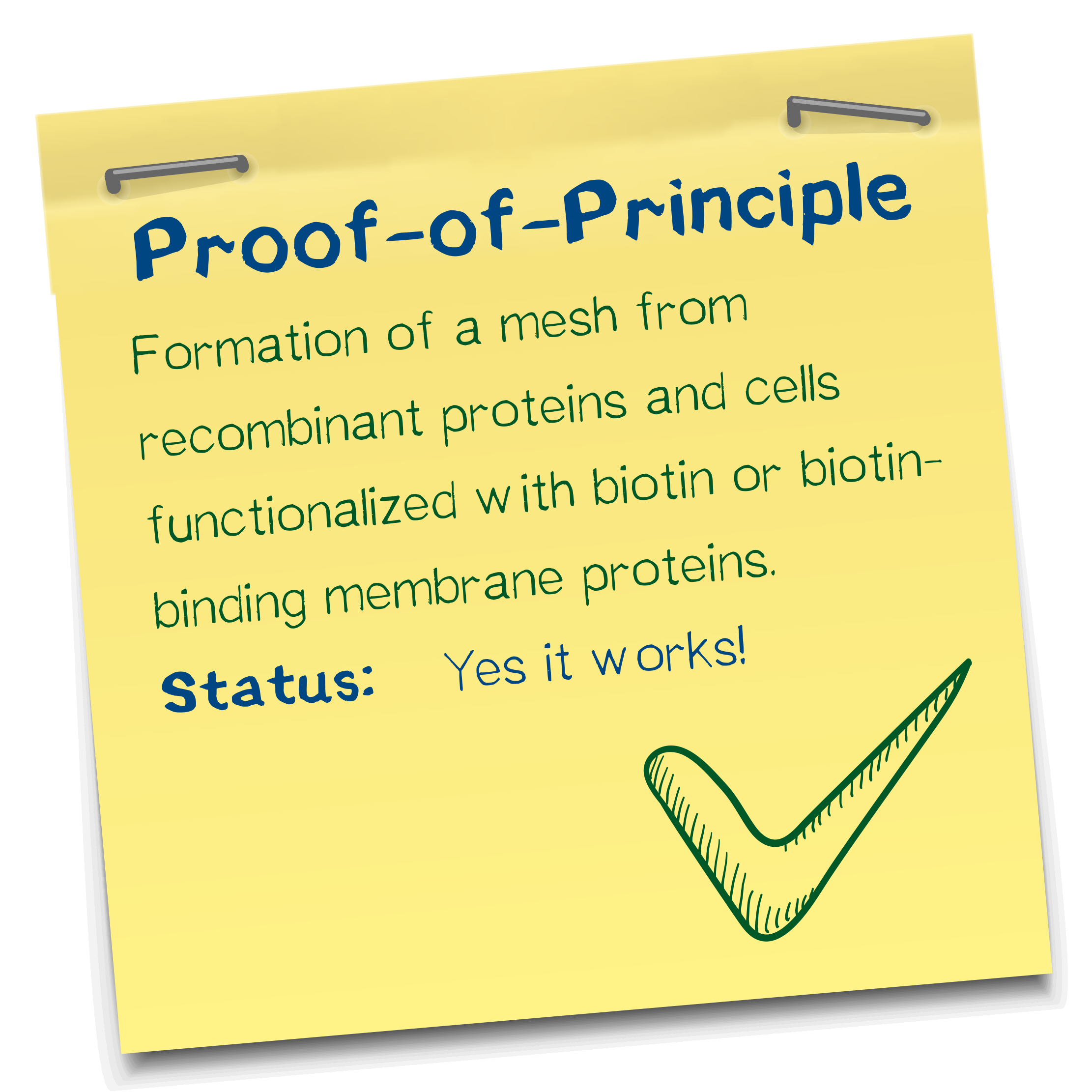
Results: Polymerization experiments
Creating the perfect mesh
In order to characterize our cells concerning their biotin binding capabilities by taking a look at their polymerization behaviour after adding them to a medium, which is rich of biotinylated protein linkers.
Since the expression of the two biotin binding receptor constructs was significantly higher than the expression of the biotin presenting one, we mainly used one of the biotin binding constructs, the scAvidin version. The main idea behind the polymerization experiments was that we wanted to find the perfect conditions for the cells to stick together, concerning the right medium, concentration and biotinylation degree of the protein linker.
The main components of all our polymerization experiments were svAvidin-construct-transfected Trex cells, biotinylated bovine serum albumine and streptavidin (or streptactin).
Our first experiments were not conducted with our printer, but under a fluorescence microscope for better analysability of the produced polymer.
The tests showed that the polymerization effect is only dependent on three things:
- cell density in the biotINK
- concentration of biotinylated BSA in the biotINK
- concentration of Streptavidin inside the printing medium
Since the addition or removal of biotinylated eGFP from the ink did not show a significant effect on then polymerization it was added to every sample for better visualization.
Experiments showed that the concentration of biotinylated BSA in our biotINK is the limiting factor for polymerization. The Higher the concentration of BSA was, the more structural integrity of the "printed" polymer could be oberved.
To see how well our printer performs with cells we wanted to test the interaction of the scAvidin transfected cells with biotinylated surfaces. For this reason we coated an ibidi-slide with poly-D Lysin for the following biotinylationwhich was then biotinylated. The cells were printed on the surface to observe, if the cells stick to the surface and are held in the right position so that the piture that is printed can be seen on the ibidi-slide. The first experiments showed that the interaction was present (Fig.x) and the results looked very satisfying.
The next step was the actual printing of our cells in the stretavidin medium to observe if the printed stucture is stable enough so that we can see the printed picture. Satisfyingly this method showed amazing results which showed us that the stuctural integrity of our polymer is high enough for printing of small 3D stuctures (Figure. Wurst)
Sticky cells in the Micromanipulator
To get a clearer view on how the cells polymerize we wanted to see the actual polymerization event and how the cells stick to each other. In order to do so we used a micromanipulator for ejection of single cells. We compared the ejection of our biotINK in Streptavidin solution with the ejection in phosphate buffered saline (negative controle) to make sure that, if polymerization occures, it is because of an specific interaction of the cells with biotinylated BSA and streptavidin.
It was very interesting to see that there really is a difference between the two experiments in a way that the biotINK, ejected in streptavidin, contrary to the one ejected in PBS, does show polymerization of cells.
References