(→Different polymers possible with biotINK polymerization) |
Javier.luna (Talk | contribs) (→Creating the perfect mesh) |
||
| Line 56: | Line 56: | ||
| − | The main components of all our polymerization experiments | + | The main components of all our polymerization experiments were svAvidin-construct-transfected Trex cells, biotinylated bovine serum albumine and streptavidin (or streptactin). |
Our first experiments were not conducted with our printer, but under a fluorescence microscope for better analysability of the produced polymer. | Our first experiments were not conducted with our printer, but under a fluorescence microscope for better analysability of the produced polymer. | ||
| − | [[Image:Muc16_Bioprinter_LogoDruck.jpeg |thumb|right|450px| '''Figure 2: | + | [[Image:Muc16_Bioprinter_LogoDruck.jpeg |thumb|right|450px| '''Figure 2:"' printing our team logo with biotin receptor expressing cells of biotinylated ibidi-slides]] |
<br> | <br> | ||
| Line 80: | Line 80: | ||
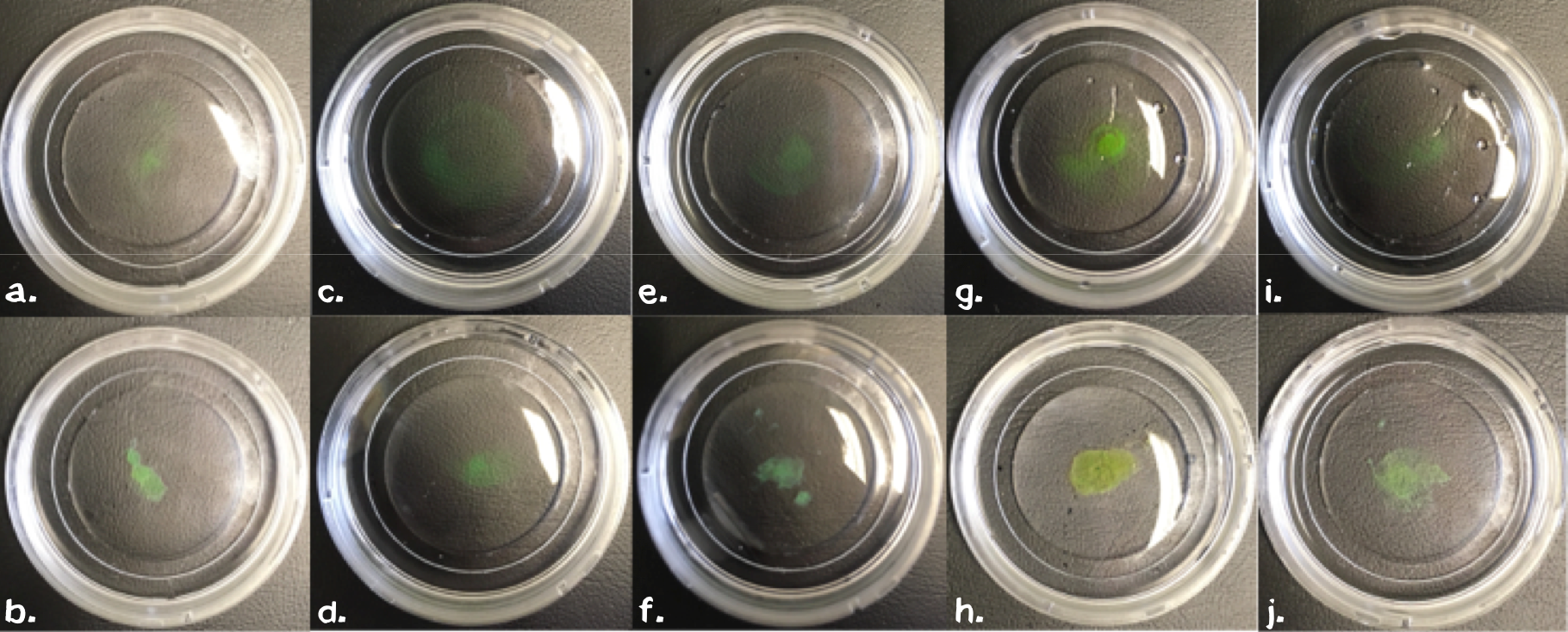
Building on this empirical approach we next evaluated the importance of these parameters for the polymer by leaving out one of each component in subsequent steps. Therefore we mixed sedimented cells, chemically biotinyltaed BSA (11 mg/ml), EGFP (3 mg/ml) or chemically biotinylated PAS-Lysine (10 mg/ml) in different settings (table X). For samples without biotinylated GFP we mixed 5 µl 1 mM non-biotinylated GFP into the samples to ensure good visualiszation. All samples were carefully swireled to ensure that agglutination is not due to an artefact. | Building on this empirical approach we next evaluated the importance of these parameters for the polymer by leaving out one of each component in subsequent steps. Therefore we mixed sedimented cells, chemically biotinyltaed BSA (11 mg/ml), EGFP (3 mg/ml) or chemically biotinylated PAS-Lysine (10 mg/ml) in different settings (table X). For samples without biotinylated GFP we mixed 5 µl 1 mM non-biotinylated GFP into the samples to ensure good visualiszation. All samples were carefully swireled to ensure that agglutination is not due to an artefact. | ||
| − | The comparission between the upper pictures in figure 4, which correspond to PBS in the reservoir and the lower ones with Streptavidin (11.4 mg/ml) in the reservoir, clearly shows that streptavidin is necessarry for our polymerization experiment. Suprisingly biotinyliated BSA or PAS-Lysine are mandatory for polymerisation. Neither cells (fig. 4d), nor GFP (fig. 4f) on its own are sufficient. By directly comparing the sample in fig. 4b with the one in fig. 4h we stated that both linker proteins either BSA or PAS-Lysine work quite good. The comparision of fig. 4b with fig. 4j shows that pre-polymerization with a small amount of Streptavidin improves the stability of the polymer, | + | The comparission between the upper pictures in figure 4, which correspond to PBS in the reservoir and the lower ones with Streptavidin (11.4 mg/ml) in the reservoir, clearly shows that streptavidin is necessarry for our polymerization experiment. Suprisingly biotinyliated BSA or PAS-Lysine are mandatory for polymerisation. Neither cells (fig. 4d), nor GFP (fig. 4f) on its own are sufficient. By directly comparing the sample in fig. 4b with the one in fig. 4h we stated that both linker proteins either BSA or PAS-Lysine work quite good. The comparision of fig. 4b with fig. 4j shows that pre-polymerization with a small amount of Streptavidin improves the stability of the polymer, nethertheless it is not mandatory. Directly printing in a Streptavidin reservoir is sufficient for polymerization. |
| − | [[Image:Muc16_testpolymerization.png |thumb|right|750px| '''Figure 4:' | + | [[Image:Muc16_testpolymerization.png |thumb|right|750px| '''Figure 4:"' Testpolymerization with different conditions]] |
| − | + | <br><br><br><br><br><br> | |
| − | All in all we developed a recipe for the „perfect | + | All in all we developed a recipe for the „perfect saucage“ we called it: |
• 6/10 volume parts biotinylated BSA, 11 mg/ml or biotinylated PAS-Lysin 10 mg/ml | • 6/10 volume parts biotinylated BSA, 11 mg/ml or biotinylated PAS-Lysin 10 mg/ml | ||
• 0.5/10 volume parts biotinylated GFP | • 0.5/10 volume parts biotinylated GFP | ||
| Line 89: | Line 89: | ||
| − | Before filling it into our printer we did some experiments to prove the stability and ability to print in a precise manner. We were absolutely thrilled by the stability and integrity of our polymer. In fig. X we pipetted the recipe given above (with | + | Before filling it into our printer we did some experiments to prove the stability and ability to print in a precise manner. We were absolutely thrilled by the stability and integrity of our polymer. In fig. X we pipetted the recipe given above (with trypane blue for visualisation) into a Streptavidin solution of 11.4 mg/ml and were able to paint with our protein-cell-polymer. In the video above we shaked the polymer without any negative influence on the stability. |
| − | Next we asked wether we were just polymerizing protein or if our receptor construct is functional and our cells are „sticky“. Therefore we needed to see that the avidinylated cells attach to the biotinylated GFP/BSA polymer. We drew some lines of our „perfect sausage“ recipe into Streptavidin solution and watched the polymer under the microscope. Here were saw a well structured shape with cells in it (fig. | + | |
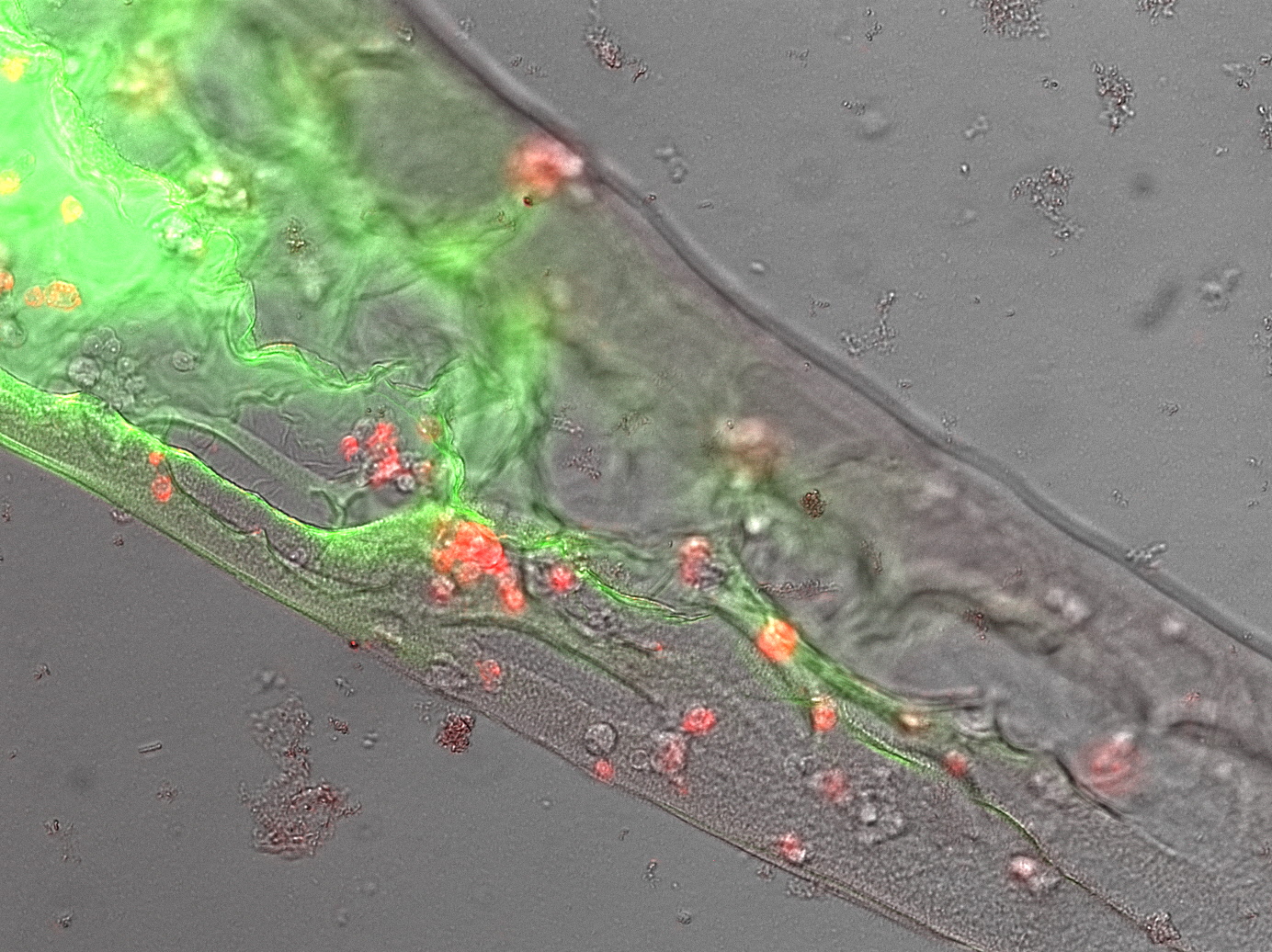
| + | [[Image:Muc16_saussage2.jpg |frameless|right|450px| '''Figure 3:"' Our genetically engineered scAvidin cells in a GFP/BSA protein-polymer]] | ||
| + | |||
| + | Next we asked wether we were just polymerizing protein or if our receptor construct is functional and our cells are „sticky“. Therefore we needed to see that the avidinylated cells attach to the biotinylated GFP/BSA polymer. We drew some lines of our „perfect sausage“ recipe into Streptavidin solution and watched the polymer under the microscope. Here were saw a well structured shape with cells in it (fig. 3). Upon heavily shaking the microscopy slide the polyer is shaking from side to side but not breaking and most interestingly: the cells are shaking with the polymer but not dropping out of it. | ||
Finally we managed to transfer all these experiments into practice: we printed the TUM logo and our own logo with a ink made of biotinylated proteins and cells! | Finally we managed to transfer all these experiments into practice: we printed the TUM logo and our own logo with a ink made of biotinylated proteins and cells! | ||
| − | [[Image:Muc16_Bioprinter_TUMDruck.jpeg |frameless|central|450px| '''Figure 5: | + | [[Image:Muc16_Bioprinter_TUMDruck.jpeg |frameless|central|450px| '''Figure 5:"' Look at what we have got: Our university logo printed with biotin receptor expressing cells of biotinylated ibidi-slides]] |
Revision as of 03:52, 20 October 2016
Design: Why we chose the Avidin & Biotin affinity pair
We have long discussed what kind of biochemical interaction we could use to have a rapid and high-affinity interaction of the components. Factors that influenced our choice were: affinity (KD), association rate (kon) as well as the number of binding sites we can have on a recombinant protein as well as the yield it gives when produced recombinantly. The comparison with some other possible affinity-pairs is shown in the following table.
| Affinity pair | Kind of interaction | Dissociation constant | Association rate |
| Avidin - Biotin | non covalent | 10-15M [1] | 55 440 |
| SpyCatcher - SpyTag | covalent | not applicable | far too slow for rapid polymerization |
| StrepTactin - Strep-tag II | non covalent | ~10-6M[2] | affinity is too low for a constant interaction |
Multivalency and avidity as major principles for polymerization
A nice point about the biotin - Avidin affinity pair is that biotin groups can be conjugated to any protein of choice that contains surface exposed amino groups. In the next line we have visualized amino groups in two proteins that we have used in our project as they are available in big amounts: enhanced Green Fluorescent Protein (eGFP, left) and Bovine Serum Albumine (BSA, right). The high number of 15 to 30 surface exposed amino groups in these proteins is a valuable advantage of this biotinylation approach as it results in highly biotinylated proteins as we also produced them in our "Proteins" sub-project. As we also synthesized the Biotin-NHS compound ourselves in our "Linker Chemistry" sub-project the supply with affordable material is also secured. Thus all components for this kind of polymerization were made accessible to the iGEM community.
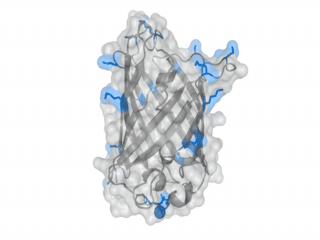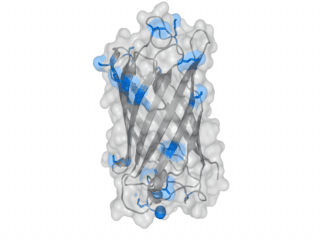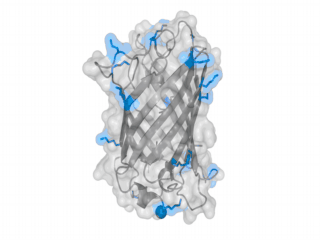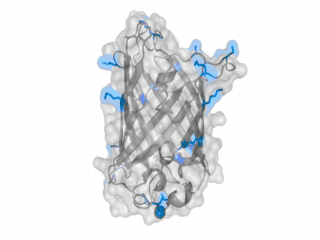

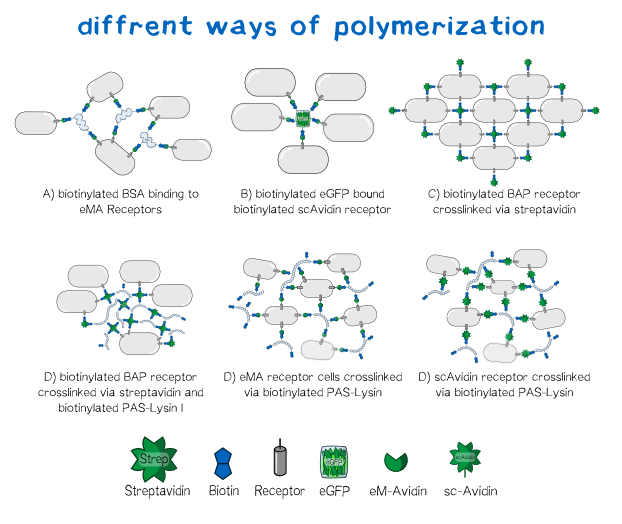
Different polymers possible with biotINK polymerization
Since the basis of our polymerization reaction is the interaction of Streptavidin and its binding partner Biotin [3] we used biotinylated polymers to convey the polymerization between the cells. As one alternative for a linker molecule we biotinylated PAS-Lysin, which is an aminoacid sequence consisting of Prolin-Alanin-Serin repeats with a Lysin at about every 20th position [4] . As part of our bioink the polymer's biotin binds to the single chain Avidin, enhanced monomeric Avidin or respectively Streptavidin and is able to crosslink the cells. Additionally we tested other molecules as linkers like bovine serum albumin or eGFP to visualize the polymerization under the microscope. The principle in this case is the same as with PAS-Lysin, though the latter is the more promising linker for its large hydrodynamic volume,the presence of 20 Lysins for biotinylation, its high solubility and lack a folded secondary structure. [5] With a variable composition of the the bioink leading to customizable results in cell aggregation the DIY principle of BiotINK is emphasized. Apart from the linker variablity our BiotINK approach offers three different possible receptors expressed on the cell surface as key points for the polymerization. Each receptor binds biotin in a more or less different. In case of scAvidin and eMA these binding are cooperativly based on the streptavidin-biotin connection. The BAP-receptor on the other hand is biotinylated via the co-expressed BirA and can be bound to Streptavidin available in the reservoir solution. In this case streptavidin itself can again bind to linkers or other biotinylated BAP-receptor cells.
All of the approaches pictured below should lead to a close mesh in which the cells are immobilized in such way that they can easily grow together and form three dimensional tissue.
Results: Polymerization experiments
Creating the perfect mesh
In order to characterize our cells concerning their biotin binding capabilities by taking a look at their polymerization behaviour after adding them to a medium, which is rich of biotinylated protein linkers.
Since the expression of the two biotin binding receptor constructs was significantly higher than the expression of the biotin presenting one, we mainly used one of the biotin binding constructs, the scAvidin version. The main idea behind the polymerization experiments was that we wanted to find the perfect conditions for the cells to stick together, concerning the right medium, concentration and biotinylation degree of the protein linker.
The main components of all our polymerization experiments were svAvidin-construct-transfected Trex cells, biotinylated bovine serum albumine and streptavidin (or streptactin).
Our first experiments were not conducted with our printer, but under a fluorescence microscope for better analysability of the produced polymer.
The tests showed that the polymerization effect is only dependent on three things:
- Cell density in the biotINK.
- Concentration of biotinylated BSA in the biotINK.
- Concentration of Streptavidin inside the printing medium.
Since the addition or removal of biotinylated eGFP from the ink did not show a significant effect on then polymerization it was added to every sample for better visualization.
Experiments showed that the concentration of biotinylated BSA in our biotINK is the limiting factor for polymerization. The Higher the concentration of BSA was, the more structural integrity of the "printed" polymer could be oberved.
The next step was the actual printing of our cells in the stretavidin medium to observe if the printed stucture is stable enough so that we can see the printed picture.
For approaching the optimal polymerisation conditions we tried lots of different combinations and concentrations of linker moleclues, cells and reservoir conditions. Based on this empirical approach we identified the critial parameters on the macroscopic scale.
- Chemically biotinylated BSA, EGFP and PAS-Lysine were the most promising linker molecules
- Our cells carrying the scAvidin were the strongest binding ones.
Building on this empirical approach we next evaluated the importance of these parameters for the polymer by leaving out one of each component in subsequent steps. Therefore we mixed sedimented cells, chemically biotinyltaed BSA (11 mg/ml), EGFP (3 mg/ml) or chemically biotinylated PAS-Lysine (10 mg/ml) in different settings (table X). For samples without biotinylated GFP we mixed 5 µl 1 mM non-biotinylated GFP into the samples to ensure good visualiszation. All samples were carefully swireled to ensure that agglutination is not due to an artefact.
The comparission between the upper pictures in figure 4, which correspond to PBS in the reservoir and the lower ones with Streptavidin (11.4 mg/ml) in the reservoir, clearly shows that streptavidin is necessarry for our polymerization experiment. Suprisingly biotinyliated BSA or PAS-Lysine are mandatory for polymerisation. Neither cells (fig. 4d), nor GFP (fig. 4f) on its own are sufficient. By directly comparing the sample in fig. 4b with the one in fig. 4h we stated that both linker proteins either BSA or PAS-Lysine work quite good. The comparision of fig. 4b with fig. 4j shows that pre-polymerization with a small amount of Streptavidin improves the stability of the polymer, nethertheless it is not mandatory. Directly printing in a Streptavidin reservoir is sufficient for polymerization.
All in all we developed a recipe for the „perfect saucage“ we called it:
• 6/10 volume parts biotinylated BSA, 11 mg/ml or biotinylated PAS-Lysin 10 mg/ml
• 0.5/10 volume parts biotinylated GFP
• 4/10 volume parts gravity sedimented scAvidin-cells
Before filling it into our printer we did some experiments to prove the stability and ability to print in a precise manner. We were absolutely thrilled by the stability and integrity of our polymer. In fig. X we pipetted the recipe given above (with trypane blue for visualisation) into a Streptavidin solution of 11.4 mg/ml and were able to paint with our protein-cell-polymer. In the video above we shaked the polymer without any negative influence on the stability.
Next we asked wether we were just polymerizing protein or if our receptor construct is functional and our cells are „sticky“. Therefore we needed to see that the avidinylated cells attach to the biotinylated GFP/BSA polymer. We drew some lines of our „perfect sausage“ recipe into Streptavidin solution and watched the polymer under the microscope. Here were saw a well structured shape with cells in it (fig. 3). Upon heavily shaking the microscopy slide the polyer is shaking from side to side but not breaking and most interestingly: the cells are shaking with the polymer but not dropping out of it.
Finally we managed to transfer all these experiments into practice: we printed the TUM logo and our own logo with a ink made of biotinylated proteins and cells!
Discussion:
GFP is carrying 13 surface lysine that can be biotinylated nethertheless it seems to need the hydrophobicity of PAS or BSA for efficient polymerisation. The need for a protein matrix that binds the avidinylated cells might be a technical one, because the cells are too far away from each other to build any contacts when leaving the syringe. Further optimization needs to be done to reach a sufficient cell density and flow-through speed.
Sticky cells in the Micromanipulator
To get a clearer view on how the cells polymerize we wanted to see the actual polymerization event and how the cells stick to each other. In order to do so we used a micromanipulator for ejection of single cells. We compared the ejection of our biotINK in Streptavidin solution with the ejection in phosphate buffered saline (negative control) to make sure that, if polymerization occurs, it is because of an specific interaction of the cells with biotinylated BSA and streptavidin.
It was very interesting to see that there really is a difference between the two experiments in a way that the biotINK, ejected in streptavidin, contrary to the one ejected in PBS, does show polymerization of cells.
References
- ↑ Green, N. M. (1963). Avidin. 1. The use of [14C] biotin for kinetic studies and for assay. Biochemical Journal, 89(3), 585.
- ↑ http://www.iba-lifesciences.com/
- ↑ Korndörfer, I. P., & Skerra, A. (2002). Improved affinity of engineered streptavidin for the Strep‐tag II peptide is due to a fixed open conformation of the lid‐like loop at the binding site. Protein science, 11(4), 883-893.
- ↑ Breibeck, J., Serafin, A., Reichert, A., Maier, S., Küster, B., & Skerra, A. (2014). PAS-cal: a Generic Recombinant Peptide Calibration Standard for Mass Spectrometry. Journal of The American Society for Mass Spectrometry, 25(8), 1489-1497.
- ↑ Schlapschy, M., Binder, U., Börger, C., Theobald, I., Wachinger, K., Kisling, S., ... & Skerra, A. (2013). PASylation: a biological alternative to PEGylation for extending the plasma half-life of pharmaceutically active proteins. Protein Engineering Design and Selection, 26(8), 489-501.