Eucaryotic receptor constructs
This is a page that is required for the evaluation of the best measurement category. Here we present the measurements of our membrane protein toolbox that allows surface-display of functional protein domains on prokaryotic as well as eukaryotic cells. If you want to read the whole story including the design of our modular receptors, please go to our Receptor page.
Subcellular protein localization using fluorescence microscopy
For better traceabilty we supplied all of our receptor constructs with reporter genes like mRuby and/or Nanoluciferase.
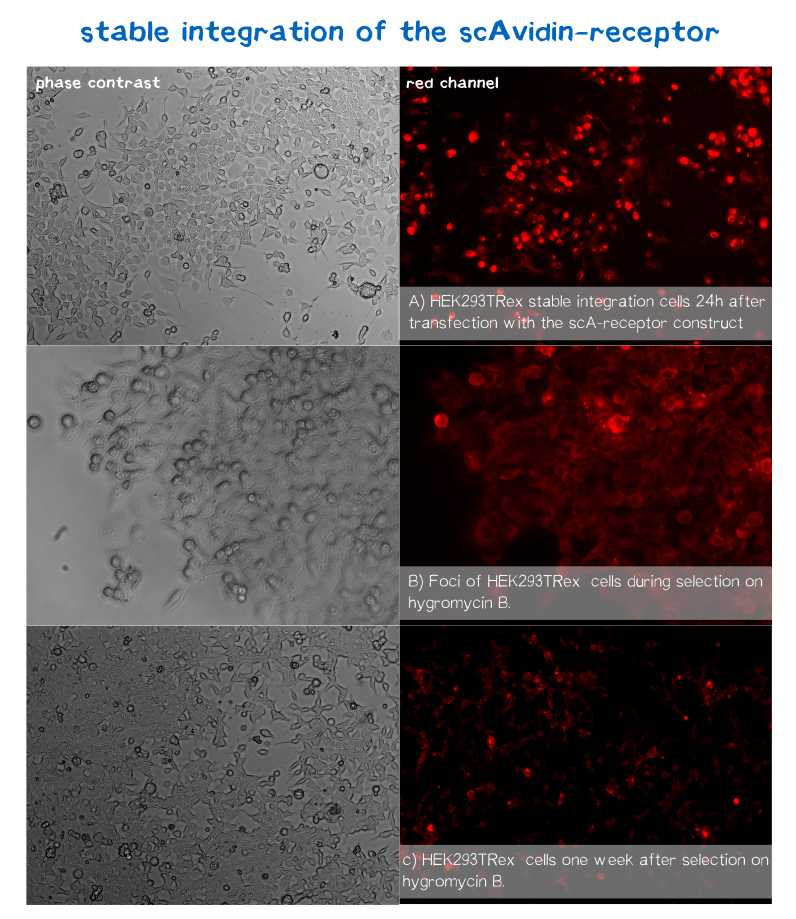
mRuby is located on the C-terminus respectively the cytosolic end of our transmembrane receptors right before the Strep-tag. Cells that express and fold the protein properly should give a signal of 605 nm (red) when exited at around 558 nm (rfp channel).[1] We tested our contructs three diffrent cell types: HEK293T adherent cells that were stably transfected, HEK293E suspension cells that were semi-stably transfected and N2a cells, a mouse neuroblastoma cell line, that we transiently transfected.
As you can see on the images the signal developed over time. In the beginning, before selection, the protein was visible in little dots and therefor seemed to be localized more in subcellular oragnelles than in the membrane. During selection on hygromycin B resistency most of the cells died, leaving some resistant cells behind, which over time grew to bigger foci consisting of stably transfected cells only (See Fig. B). For the BAP-receptor there was hardly to no fluorescence detectable neither in semi-stable nor in the stable transfection approach. But in latter case the cells still grew on hygromycin B and formed foci, which leads us to the conclusion that chromosomal intergration was susccesful (as already shown via PCR) hence there must be some problem in later steps of transcription, translation or protein maturation. After 2 weeks of selection on hygromycin B the foci were harvested and expanded. The cells seemed to recover fast and moreover the red dots nearly disappeared leaving only the membrane of the scA-receptor cells illuminated by mRuby.
High resolution confocal fluorescence microscopy
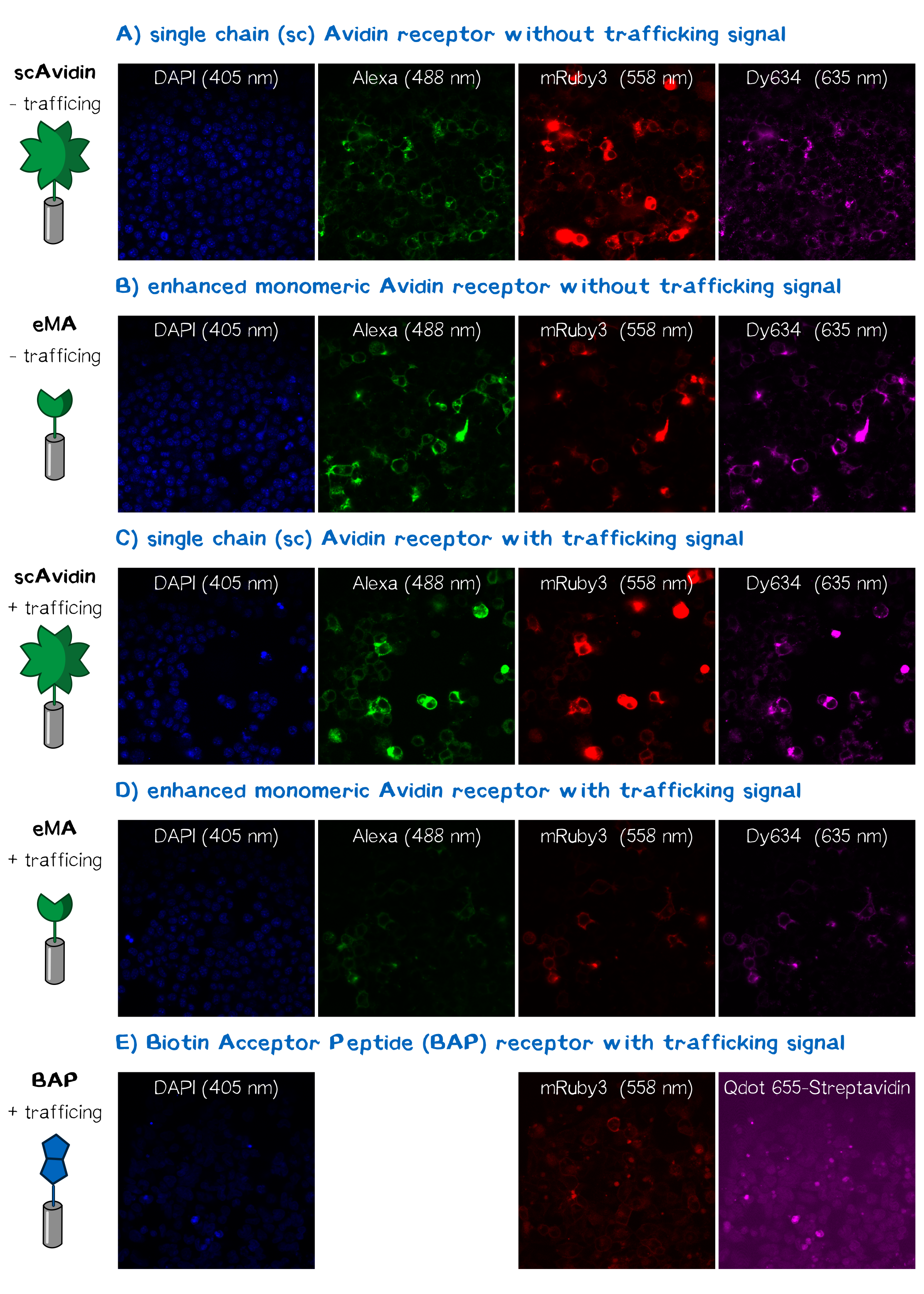
In order to obtain high resolution confocal fluorescence images from our receptor constructs we transfected the Neuro-2a (N2a) mouse neuroblastoma cells line with the respective plasmids. Shortly before the end of the project Christoph came up with the idea to include a trafficking signal to the C-terminus of the receptor proteins that is described to increase the surface presentation of receptors by sorting them intracellularly towards the cell membrane. The sequence of this trafficking signal is DFFdDFFJDFJDKJFKFDJFD (aagagcaggatcaccagcgagggcgagtacatccccctgg accagatcgacatcaacgtgGGCGGCttctgctacgagaacgaggtgtaa)[2] and we have inserted it into the assembled BioBrick device using PCR cloning. After the transfection of the receptor plasmids cells were stained with the following reagents: Hoechst dye for the nuclear staining at 405 nm; Alexa488-bioting to stain biotin binding proteins; mRuby3 which is a part of the receptor and is expected to show a membrane localization and Dy634-antiA3C5-Fab. The staining was followed by three washing steps with PBS. Following this staining procedure the cells were mounted on a confocal fluorescence microscope (Leica). The results for each fluorescence channel are given in the following figure (see Fig. ???).
Quantification of functionalized membrane proteins using flow cytometry
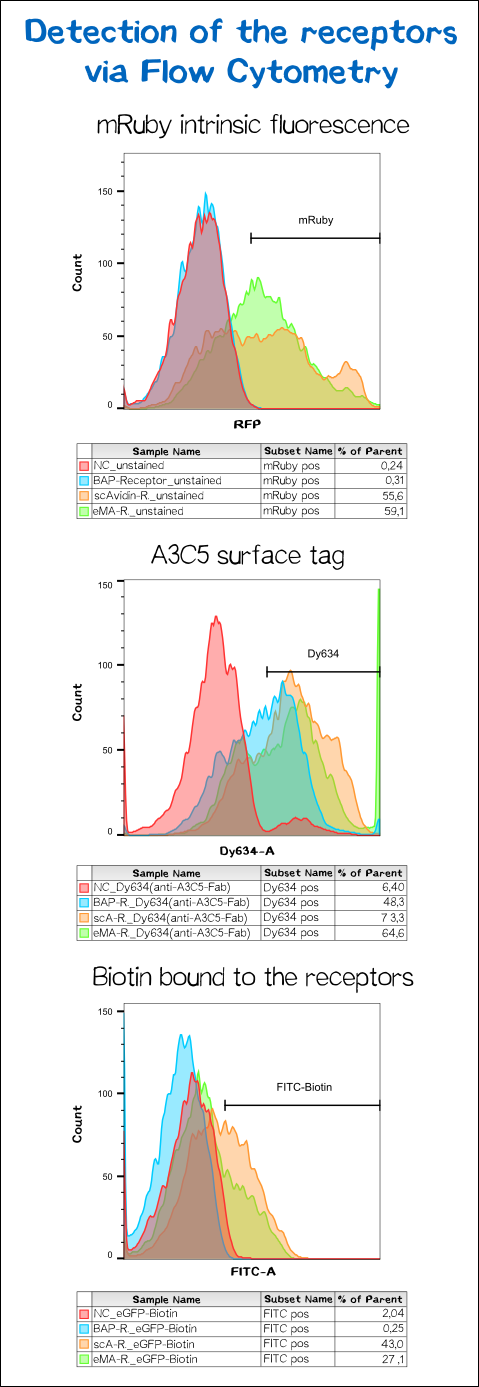
To make sure the receptor is properly presented on the surface of the cells and to quantify the yield of stably transfected cells we used flow cytometry analysis.
The results presented in Fig. 11 show a positive signal in the rfp channel for the scAvidin and the eMA receptor bearing cells. As already observed in microscopy mRuby-fluorescence could not be detected for the BAP-receptor cells. But surprisingly about 40% where positive for Dy634, meaning that the A3C5 tag, against which the fluorophor labeled Fab is dierectet, must be present on the cell surface. This results suggest that there might be a problem about the folding of the mRuby beta-barrel leading to a loss of fluorescent activity.
For scA and eMA cells Dy634 signal has been positive albeit not as high as expected.
Another stain was the eGFP coupled with biotin giving a positive signal for scA and eMA to which the biotin binds. For BAP cells there was no signal since this receptor, is biotinylated by the BirA and doesn't cooperatively bind the Biotin. Moreover the biotin snd the dye are coupled via the C-terminal tail of the biotin leaving no contact point for the BirA to ligate the eGFP-biotin to the receptor's acceptor peptide.
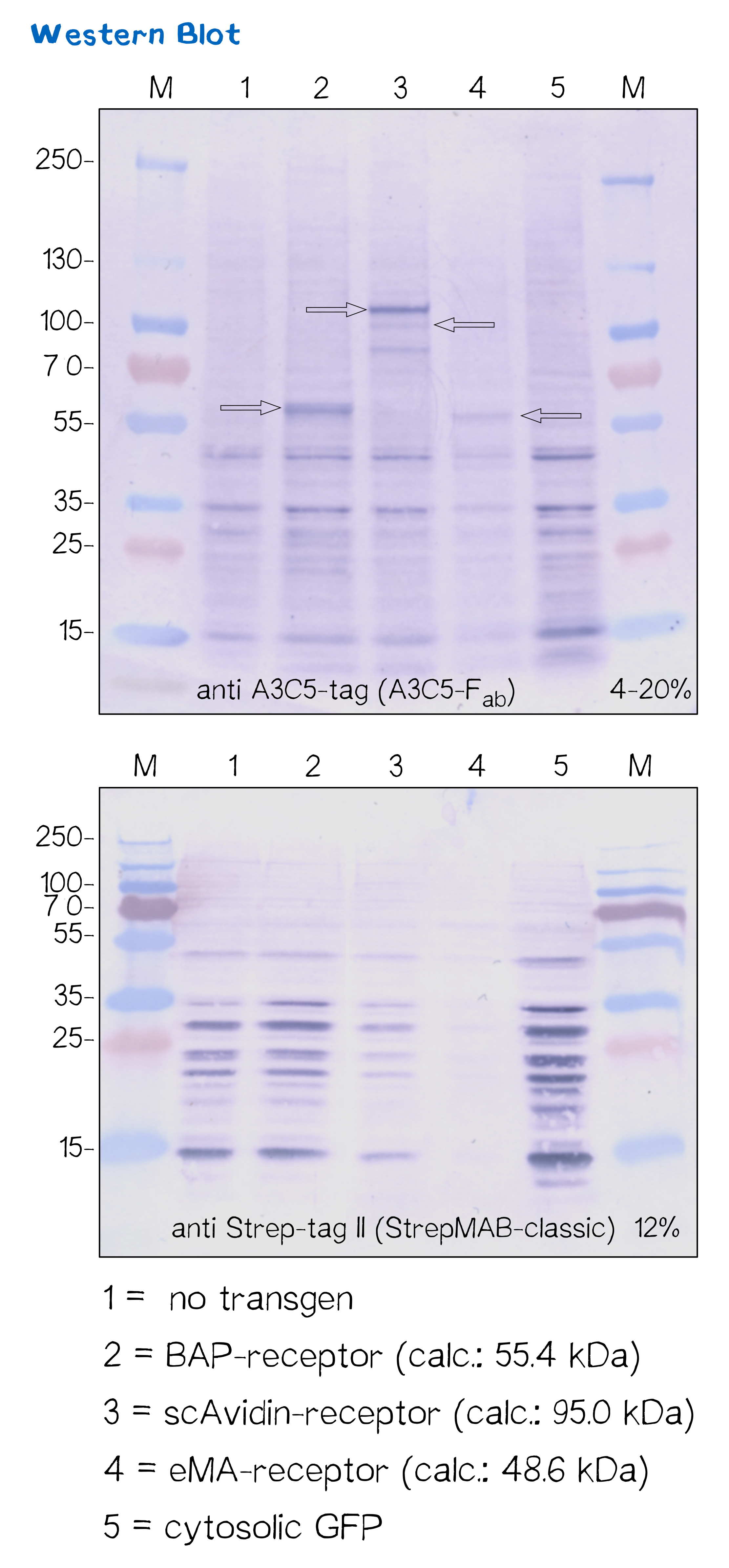
Immunochemical detecton of functionalized membrane protein using Western Blot
Another way to verify the succesful transfection and protein expression is given by the two tags (A3C5 and Strep-tag II) that are included in the receptor design, which allow detection via Western Blot analysis.
After transfecting mammalian cells with the receptor constructs and selecting them for 2 weeks on hygromycin, we lysed the cells and checked for the presence of Strep- and A3C5-tag via western blot using primary antibodies (or a Fab for A3C5) against the respective tags und secondary alkaline phosphatase coupled antibodies (see methods). As the figure shows, the Strep-tag wasn't detectable via WB - however, the A3C5-tag-WB shows significant singular bands at the size of our proteins in the respective samples.
This data corresponds with the flow cytometry results again showing that the BAP receptor is also present in the non-fluorescent cells.
| Receptor | BioBrick | Amino acids | Moelcular mass [Da] |
| BAP-Receptor | [http://parts.igem.org/Part:BBa_K2170000 BBa_K2170000] | 507 | 55 440 |
| eMA-Receptor | [http://parts.igem.org/Part:BBa_K2170001 BBa_K2170001] | 448 | 48 618 |
| scAvidin-Receptor | [http://parts.igem.org/Part:BBa_K2170002 BBa_K2170002] | 877 | 95 028 |
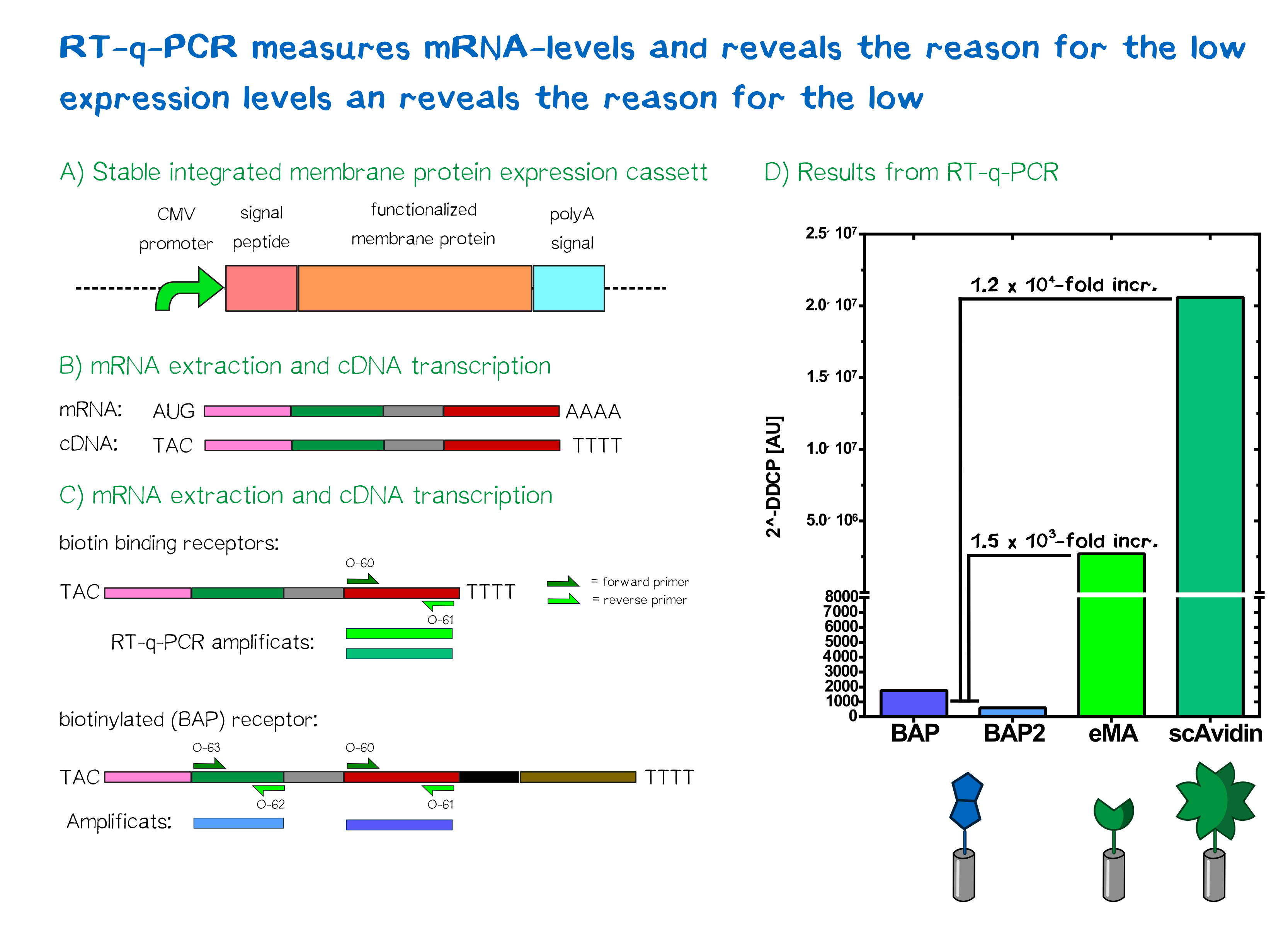
Quantification of mRNA expression levels by RT-q-PCR
The characterization of our transfected mammalian cells showed that the two receptors containing an avidin variant were expressed and translocated to the outer cell membrane. The biotin-presenting receptor containing the biotin-acceptor peptide, though, showed a very low fluorescence signal. Therefore, the success of the stable integration was tested by PCR of the genomic integration, which lead to the conclusion that the construct was transfected and integrated sucessfully, hinting at a problem downstream of the integration. In order to understand where the source of this problem was, we took a closer look at the expression levels of the different receptors by analyzing mRNA levels in comparison to each other and to the housekeeping gene (HKG) GAPDH.
For a better comparibility, we used the same amount of cells for each receptor and not only reviewed cells with stably integrated but also cells with the episomal stable receptor constucts. After extraction of the total mRNA from the cells the quality and integrity was measured by Bioanalyzer. All samples showed a RNA integrity number (RIN) above 9 which means that the total mRNA is pure and does not have any contamination. During the first step of RT-qPCR the total mRNA in converted to cDNA by a reverse transcriptase, which is then used in the actual quantitative PCR to quantify the amount of a specific mRNA was produced by the cells. For this reason two primer pairs were designed. One of them was designed to bind the exact same region in all of the different receptor mRNAs and amplify the same part of the receptor. Contrary to that the second pair was supposed to bind only in the biotin presenting receptor and amplify a part that is unique to this receptor. By this method we wanted to observe if only one part of the mRNA is unstable or if the general mRNA level of this receptor is decreased.
For the evaluation of the raw data we used the ΔΔCP method[3] in which the crossing points (CP values) of a gene of interest and of a reference gene are compared and normalized with a constantly expressed HKG, to get an idea of the relative amounts of the mRNA of interest.
In the following you can see the results of our experiments. We compared the expression levels of T-REx cells which were transfected with our different receptor constructs and with non-transfected (NT) T-REx cells.
As can be seen the ΔΔCP values of the mRNA of the eMA and scAvidin constructs are with a 1,2 x 106 fold increase for scAvidin and 1.5 x 103 fold increase for eMA, significantly higher than those of the BAP construct. The analysis of the different primer pairs in the two regions of the BAP construct also showed similar values and thus confirms the reliability of the measurement. A difference in CP value is tantamount to relative mRNA differences between cells.
Together with the result that the integration of the constuct into the genome was successful we can conclude that our first assumption, that the missing RFP signal in cells, which were transfected with the BAP construct, is caused by a reduced mRNA stability, is correct.
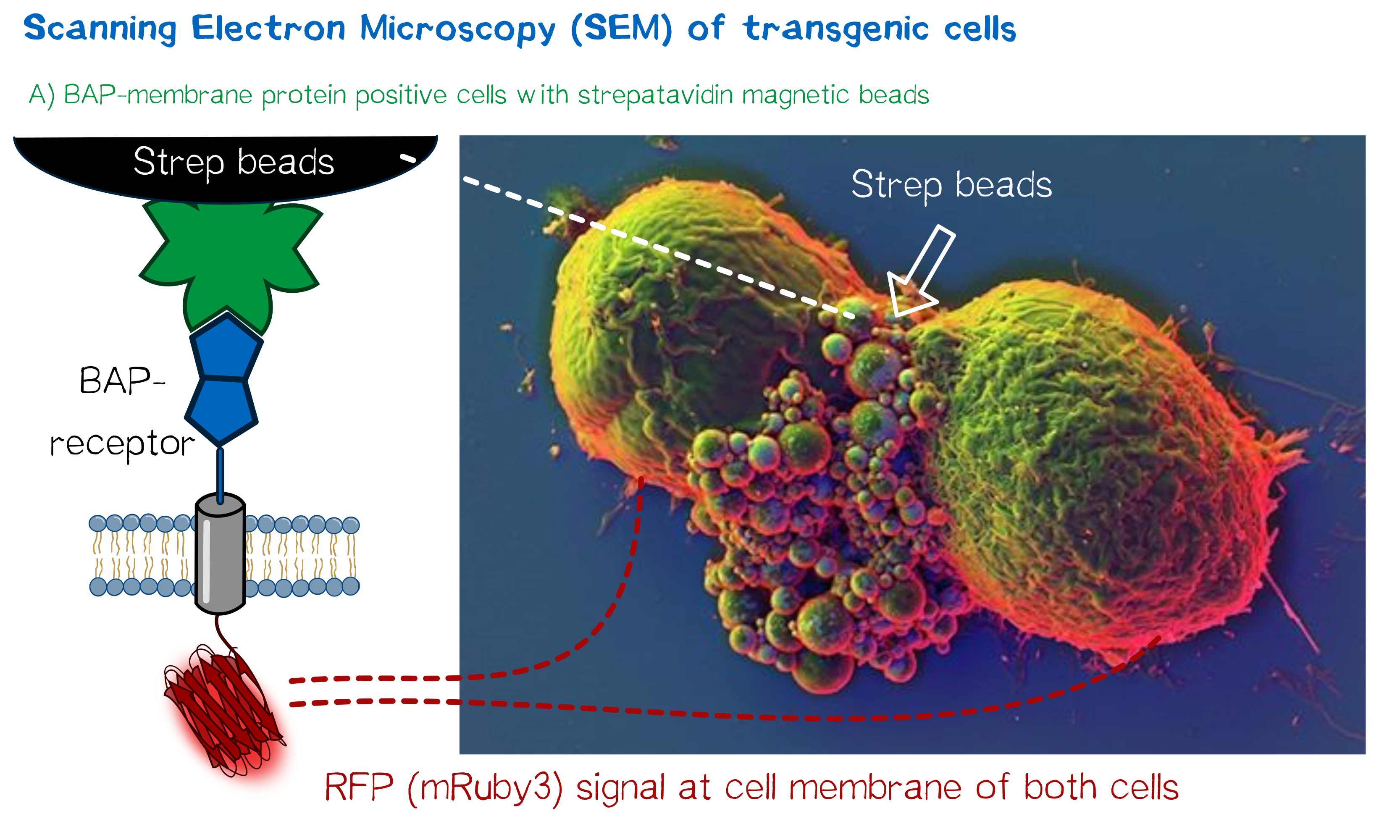
High resolution imaging using scanning electron microscopy (SEM)
As we wanted to obtain high resolution images from the transgenic cells transfected with our modular membrane proteins we contacted one of the experts on electron microscopy in Munich and luckily Prof. em. Dr. Wanner (LMU Munich) agreed to support our project by contributing scanning electron microscopic (SEM) imaging. Even better suitable for our purpose Prof. Wanner has developed a method in which he can correlate the fluorescence signal of a sample with the high resolution images obtained in SEM. As this kind of sample preparation, sample processing and imaging takes a while, we had to start 2 weeks before wiki freeze and at that point we did not have enough stably integrated T-REx 2cells, yet. Thus we used transient transfected, semi-stable MEXi HEK 293T cells that were pulled down with magnetic beads in order to increase the number of transgenic cells in the preparation and to show the specific binding of the beads to the cells. For this immunoprecipitation we incubated scAvidin-receptor and eMA-receptor cells with biotinylated magnetic beads and on the other side cells transfected with the BAP-receptor together with streptavidin-coupled magnetic beads. After the immunoprecipitation the cells were washed twice with PBS and it was visible that the portion of transgenic (red fluorescent) cells was largely increased after this procedure. The depicted SEM image shows two BAP-receptor transfected MEXi cells which have a cluster of magnetic streptavidin-conjugated magnetic beads at their cellular interface. At the border of the cells the superimposed fluorescence image of these cells shows red fluorescence located at the periphery of the cells indicating the presence of mRuby3 fluorescent protein at the cellular membrane. The green fluorescence is caused by biotinylated eGFP that was used for other preparations on the same microscopic slide and came into contact with these cells when the suspension cells were sedimented on the microscopic slide by centrifugation. Although for a reliable conclusion one would have to evaluate several such images this figure already gives a valuable evidence that the BAP-receptor is present on the cells (although perhaps at lower levels) and can bind to strepavidin that was conjugated to the beads.
Procaryotic receptor constructs
Design of an autotransporter construct for bacterial surface display
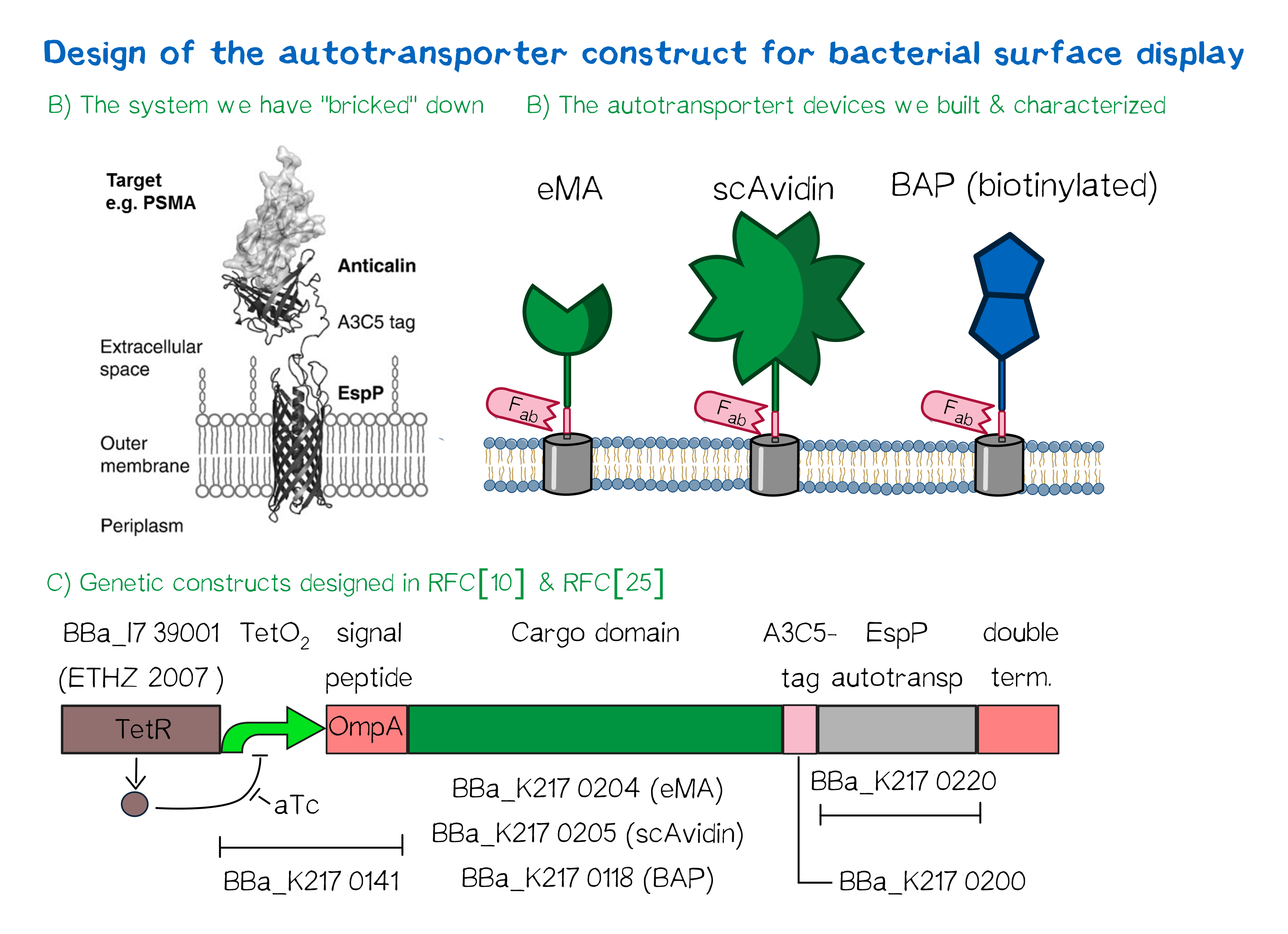
Besides the design of biotinylated and biotin-binding receptors for eukaryotic cells, we also wanted to apply our biotINK approach for prokaryotic cells. For this purpose, we designed an autotransporter device that is able to present biotinylated or biotin-binding protein domains on the surface of E. coli. Constructs for bacterial surface display are hereby already well known in the field of protein engineering of therapeutic proteins, such as antibodies. There, they are used to screen libraries of different antibodies concerning their affinity towards a given therapeutic target.
We based our design on the autotransporter EspP[4].
The autotransporter system itself is established and functional (see Fig. A[5]), and here we "bricked" it down into RFC[10] and RFC[25] BioBricks. At first we tested different inducible bacterial promoter systems (e.g. "TetR repressed GFP"; [http://parts.igem.org/Part:BBa_K577893 BBa_K577893]) that are available in the parts registry, but when we saw that two of the available systems did not work in our own hands, we decided to built the system on our own. We used the Tet-Repressor generator that was available on the distribution plates ([http://parts.igem.org/Part:BBa_I739001 BBa_I739001]) as we believe that it is very important for the standardization in SynBio to use available parts. Downstream of this repressor generator we designed and fused a BioBrick ([http://parts.igem.org/Part:BBa_K2170141 BBa_K2170141]) that encodes a double Tet-Operator (Tet-Repressor binding site together with a bacterial promoter and an signal peptide of the outer membrane protein A (OmpA[6]) from E. coli that has a 3' RFC[25] restriction site, allowing the protein fusion of proteins that are secreted into the bacterial periplasm when expressed with this BioBrick. As a protein fusion, we then assembled a protein cargo domain that is generally exchangeable for any protein that is so small in size that it can be transported through the outer membrane by the EspP autotransporter. In our project we used the same extracellular domains as for the eukaryotic receptors: enhanced monomeric Avidin (eMA; [http://parts.igem.org/Part:BBa_K2170204 BBa_K2170204]), single-chain Avidin (scAvidin; [http://parts.igem.org/Part:BBa_K2170205 BBa_K2170205]) and a composite part composed of a N-terminal Biotinylation acceptor peptide (BAP) and a C-terminal NanoLuciferase (together shown as BAP; ; [http://parts.igem.org/Part:BBa_K2170118 BBa_K2170118], see Fig. B). Further downstream our design features a A3C5-epitope tag that can be recognized by a high affinity Fab-fragment and is generally used to detect and quantify the surface display of the cargo domain[7]. Further downstream of this affinity tag we fused the BioBrick for the EspP autotransporter from E. coli[8][9] which is then followed by a bacterial terminator. For the termination we again chose a well working terminator from the distribution plate ("double terminator"; [http://parts.igem.org/Part:BBa_B0010 BBa_B0010]-[http://parts.igem.org/Part:BBa_B0012 BBa_B0012]).
But this was just the DNA-part. What is expected to happens on a protein level? The construct constantly produces Tet-repressor that binds to thet Tet-Operator and repressed the promoter activity of the autotransporter gene. As soon as the inducer anhydrotetracycline (aTc) is added to the culture, it binds to the Tet-repressor that can't bind any more to the Tet-operator and thus the expression of the autotransporter can be regulated. Although the TetR-system is known to be a tight promoter system there is always a certain background expression with most promoters. If the protein expression of the autotransporter is induced using aTc the autotransporter is transcribed and translated. Due to the bacterial signal peptide the protein is secreted into the bacterial periplasm (the space between the two bacterial membranes of gram-negative bacteria such as E. coli). When present in the bacteria periplasm the autotransporter diffused to the outer membrane and inserts into the membrane. After the integration into the outer membrane the protein cargo is transportet through the beta-barrel of the autotransporter and is pre sented on the bacterial surface. We are sure that this BioBrick will be a valuable contribution to the Parts Registry as it allows future teams to display small protein domains on the surface of gram-negative bacteria, such as E. coli.
Results: Testing the bacterial autotransporter system
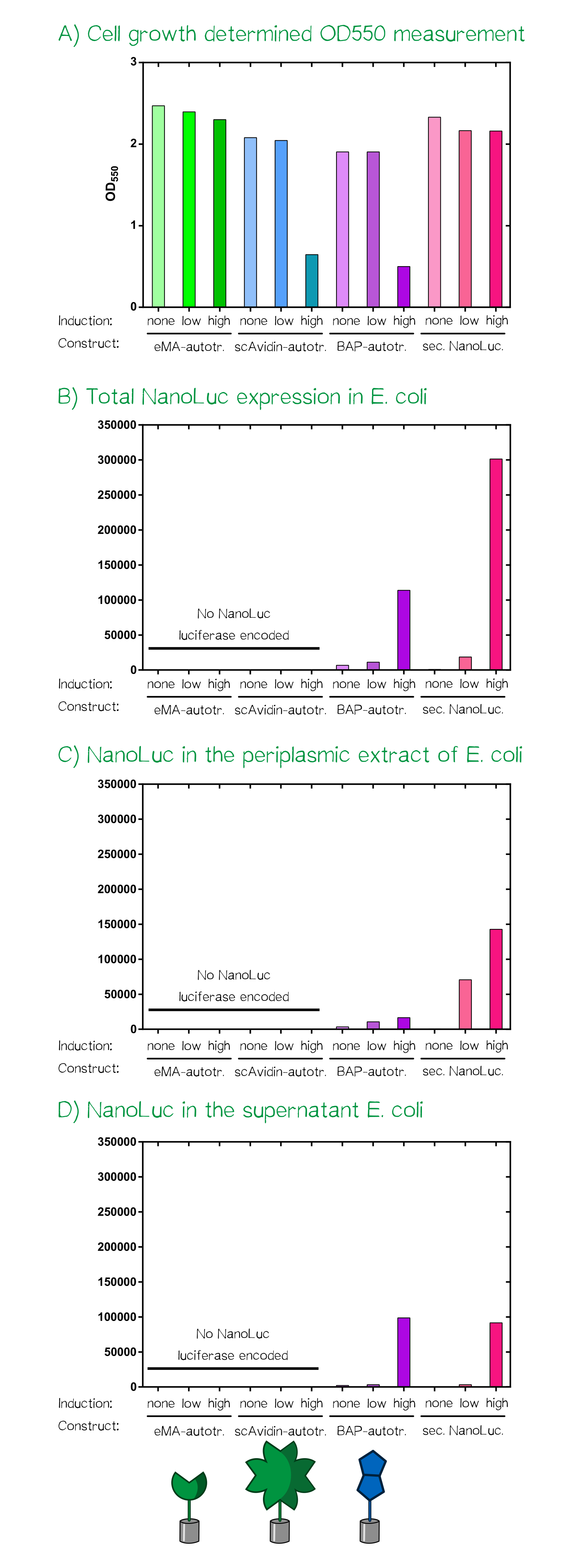
For the testing of our autotransporter constructs we transformed all three autotransporter constructs together with a secreted NanoLuc luciferase, which served as a control, into E. coli JK321[10]. Transformed cells were grown in LB-medium supplemented with chloramphenicol (pSB1C3-vector) at 30°C and 180 RPM. In the morning a stationary over night culture was used to inoculate 50 mL fresh medium with 1 mL of culture. The cultures were grown to OD550 = 0.5 and then the induction of the protein production was started using aTc: "non" = non-induced; "low" = 25 ng/mL aTc; "high" = 200 ng/mL. After induction the cultures were grown for further 5 h at the same concitions and cells were afterwards stored at 4°C until they were used for analysis or bioprinting.
After the cell harvest the optical density (OD550) was determined again as it is known that a high expression of EspP makes the outer membrane of E. coli permeable and this drastically affects cellular viability. For the scAvidin-autotransporter and the secreted BAP-NanoLuc autotransporter at high inducer concentration show clearly reduced viability as they did not show any growth after induction at OD550 = 0.5 (see Fig. A). In order to analyze the functionality of the whole design and these new BioBricks we subjected each of the twelve cultures to three different treatments and determined subsequently the amount of NanoLuc luciferase using the manufacturers substrate kit (Thanks to our sponsor Promega ;).
At first the total NanoLuc luciferase content was determined in samples that were just washed once with PBS and contained the whole bacterial cell (see Fig. B), bacterial cells that were incubated with PPA-buffer (periplasmic extraction buffer; 500 mM sucrose, 100 mM Tris/HCl pH 8.0, 1 mM EDTA) that extracts the proteins present in the periplasm (see Fig. C) and finally the culture supernatant of the cultivated bacterial cells (see Fig. D). The total NanoLuc activity is only present in bacteria that are transformed with a plasmid encoding the luciferase and there is a clear dependency of inducer concentration a detected luciferase activity. In the samples where a periplasmic extract was prepared the luciferase activity is highest in the samples of the secreted luciferase as the EspP-fused BAP-NanoLuc is attached to the cells and only proteins from lysed cells are present in the periplasmic extract. In the case of the samples where the supernatant of the cultures was analyzed the luciferase activity is present in both "high" induced samples.
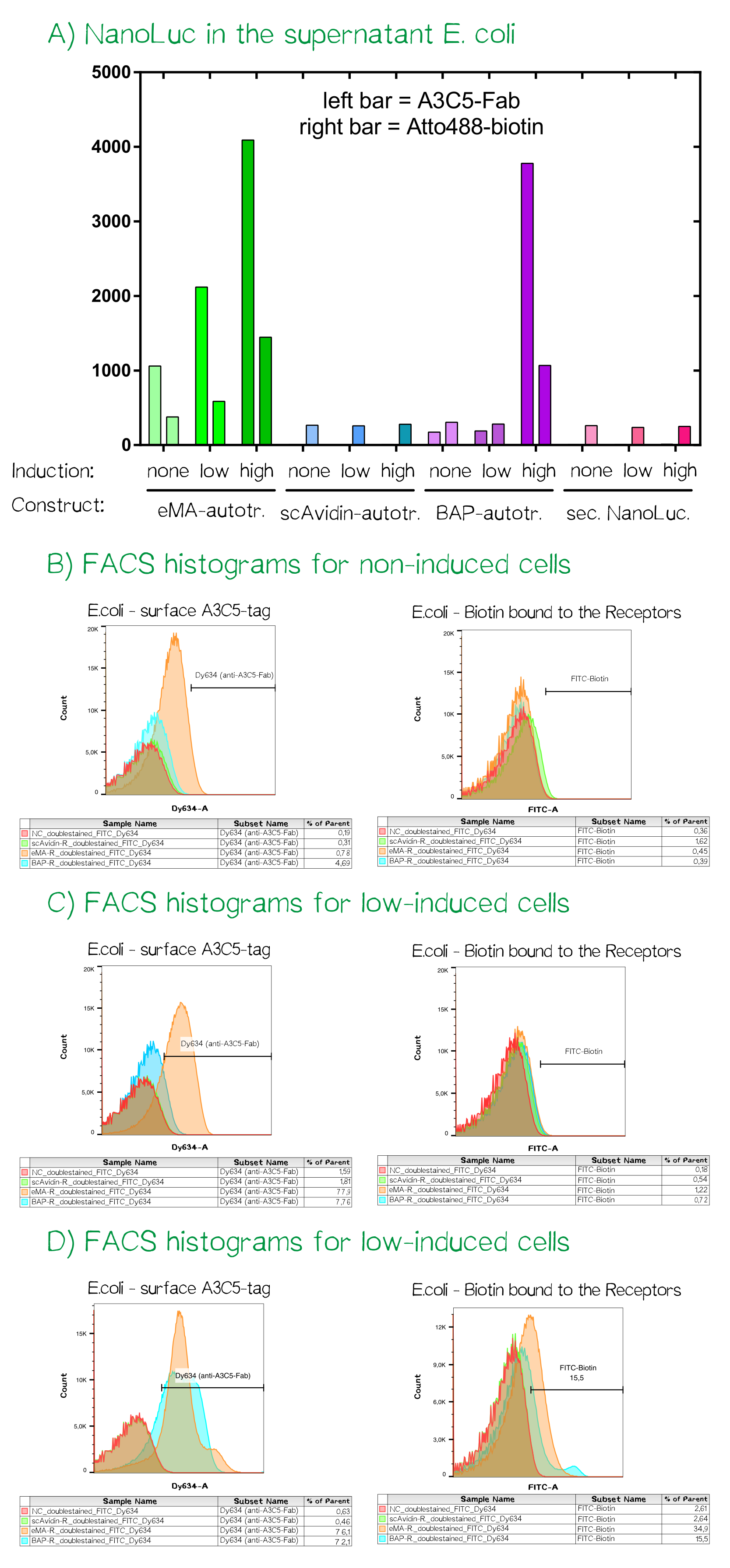
As the luciferase activity only gave an indication for the subcellular localization of the cargo-autotransporter fusion protein, we also performed flow cytometry measurements. For this purpose the same cells were stained with app. 10 µM A3C5-Fab that was coupled to the fluorescent dye Dy634-NHS and 100 µM of the chemical compound biotin-atto488. Cells were incubated for 2 h at 4°C, washed twice with PBS and analyzed using a FACSAria II flow cytometer (BD bioscience, voltages for fluorescence channels were all at 750 V).
For the channels FITC (biotin-atto488) and Dy634 (A3C5-Fab-Dy634, detected using a HeNe laser (633 nm) and a 660/20 band pass filter) the fluorescence signal was measured and mean values are given. The cells showed a homogeneious population in the FSC/SSC plot. The A3C5-Fab staining detected high signals up til a mean of over 4000 for the eMA-autotransporter dependent on the inducer concentration. For the same bacterial cells the binding of biotin-Atto488 could be confirmed as well. The single chain Avidin (scAvidin) cells showed absolutely no A3C5 signal but a low Biotin-Atto488 background signal that could be caused by biotin-importers that transport biotin into the cells or the slow diffusion into the periplasm of E. coli. In contrast the BAP-Receptor also shows a high surface presentation as seen in the A3C5 signal of nearly 4000 for the "high" induced sample. The elevated biotin-Atto488 is unexpected but could be explained by the weakened and partly permeable out membrane that does not stop the small-molecule dye from entering the periplasm.
Discussion: The autotransporter is working and can present small protein cargos on the bacterial surface
The performed experiment confirm the functionality of the device including the tetracyclin repressed promoter, the secretion into the bacterial periplasm as well as the presentation of the protein cargon on the bacterial outer membrane. In agreement with the expectation the 62 kDa big scAvidin with its extremely strong fold is too big in size for the autotransporter and can not be presented on the bacterial surface. In contrast the smaller BAP-NanoLuc (19 kDa for the NanoLuc) or the eMA (15 kDa) both can be efficiently presented on the surface. The enzymatic biotinylation of the Biotin Acceptor Peptide (BAP) using recombinant BirA biotin ligase is well established but could not be show before the wiki freeze. In Summary the whole device works great but there seems to be a limitation concerning the size of proteindomains that can be presented with the EspP autotransporter. We are curious to see what future iGEM teams will use the EspP Autotransporter in the future.
References
- ↑ Kredel, S., Oswald, F., Nienhaus, K., Deuschle, K., Röcker, C., Wolff, M., ... & Wiedenmann, J. (2009). mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PloS one, 4(2), e4391.
- ↑ Gradinaru, Viviana, et al. "Molecular and cellular approaches for diversifying and extending optogenetics." Cell 141.1 (2010): 154-165.
- ↑ Livak, Kenneth J., and Thomas D. Schmittgen. "Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method." methods 25.4 (2001): 402-408.
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Ghrayeb, J., Kimura, H., Takahara, M., Hsiung, H., Masui, Y., & Inouye, M. (1984). Secretion cloning vectors in Escherichia coli. The EMBO journal, 3(10), 2437.
- ↑ Costa, J., Grabenhorst, E., Nimtz, M., & Conradt, H. S. (1997). Stable expression of the Golgi form and secretory variants of human fucosyltransferase III from BHK-21 cells Purification and characterization of an engineered truncated form from the culture medium. Journal of Biological Chemistry, 272(17), 11613-11621.
- ↑ Barnard, T. J., Dautin, N., Lukacik, P., Bernstein, H. D., & Buchanan, S. K. (2007). Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nature structural & molecular biology, 14(12), 1214-1220.
- ↑ Skillman, K. M., Barnard, T. J., Peterson, J. H., Ghirlando, R., & Bernstein, H. D. (2005). Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Molecular microbiology, 58(4), 945-958.
- ↑ Jose, J., Krämer, J., Klauser, T., Pohlner, J., & Meyer, T. F. (1996). Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga β autotransporter pathway. Gene, 178(1), 107-110.