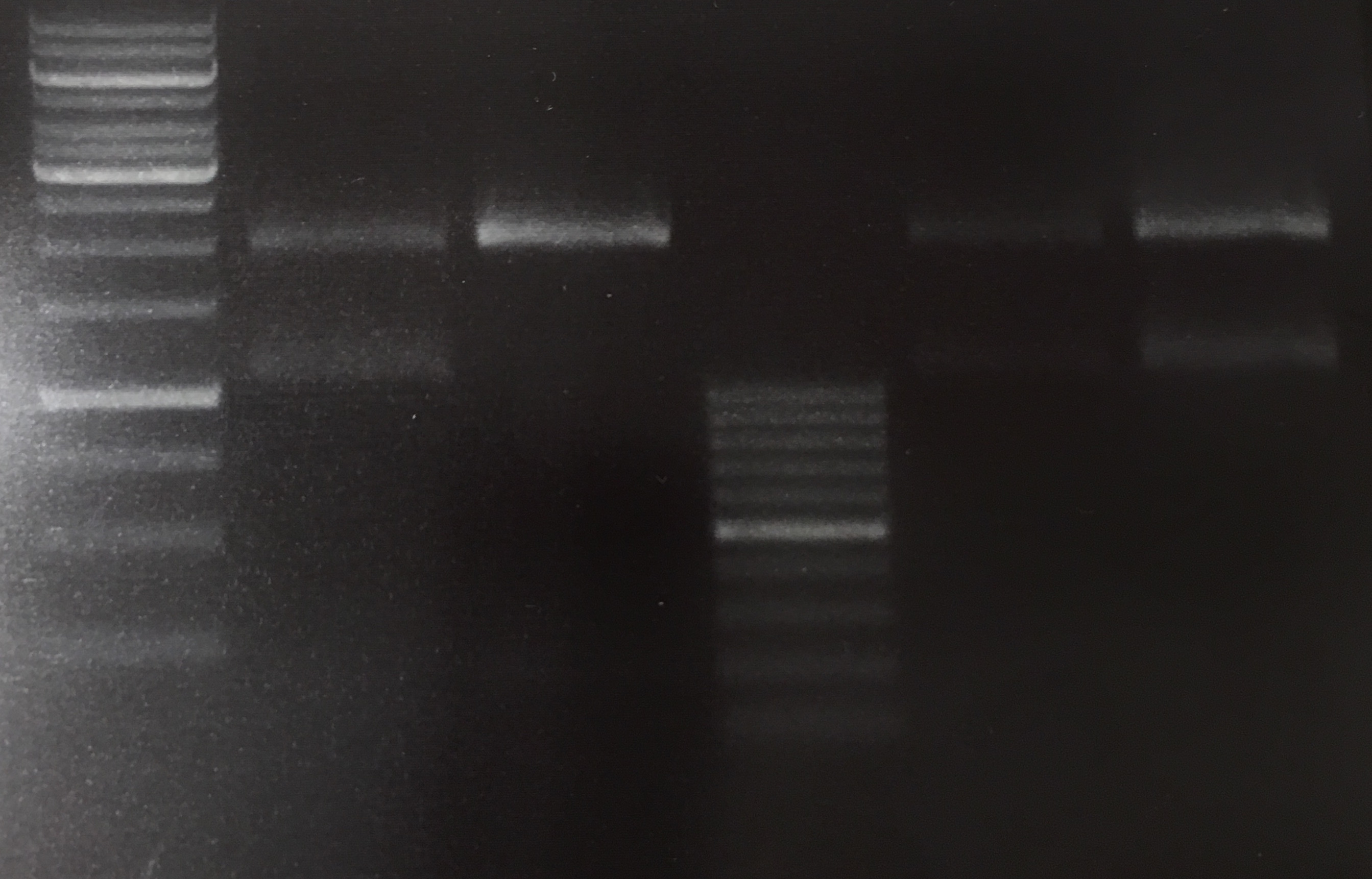
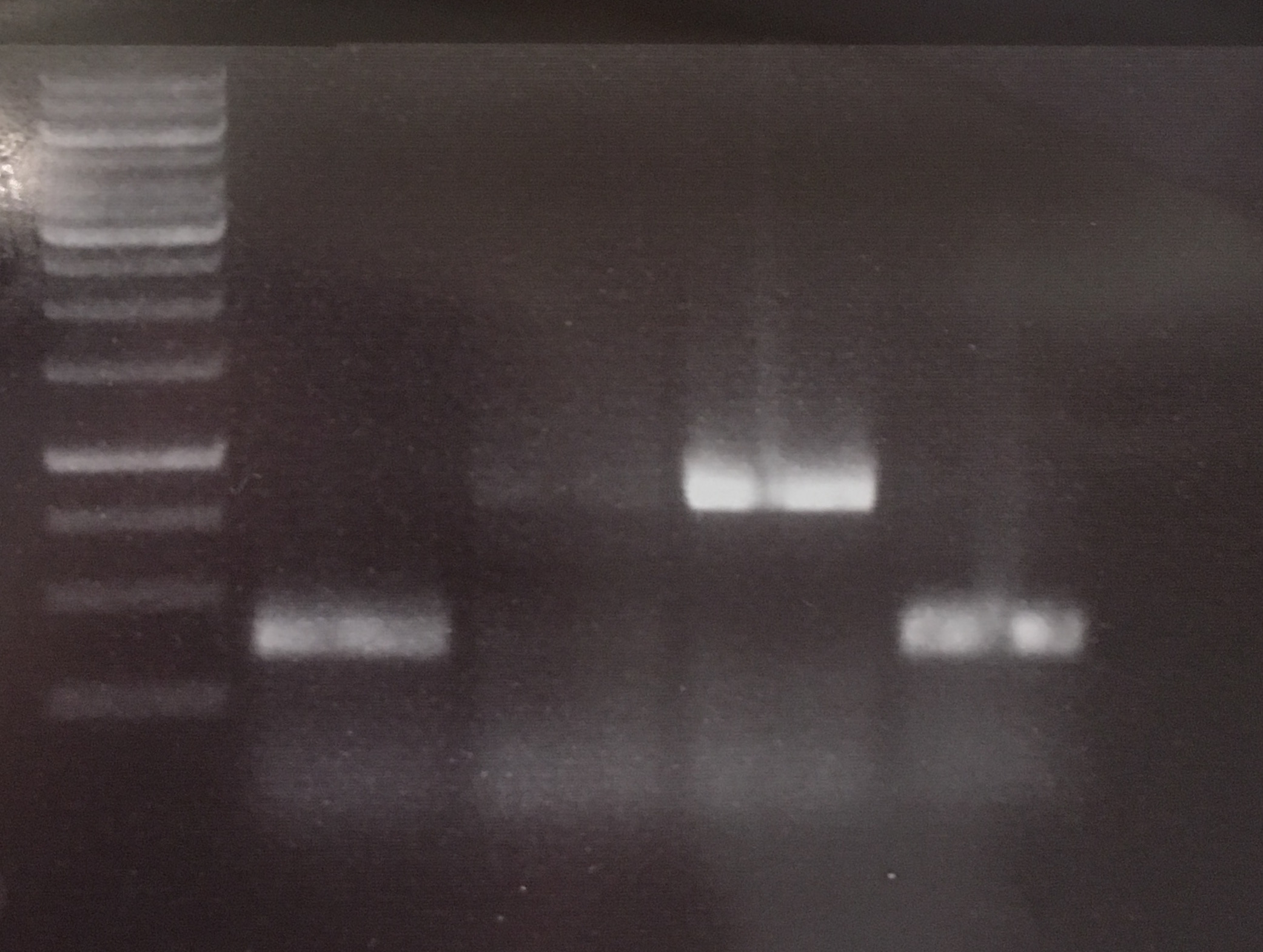
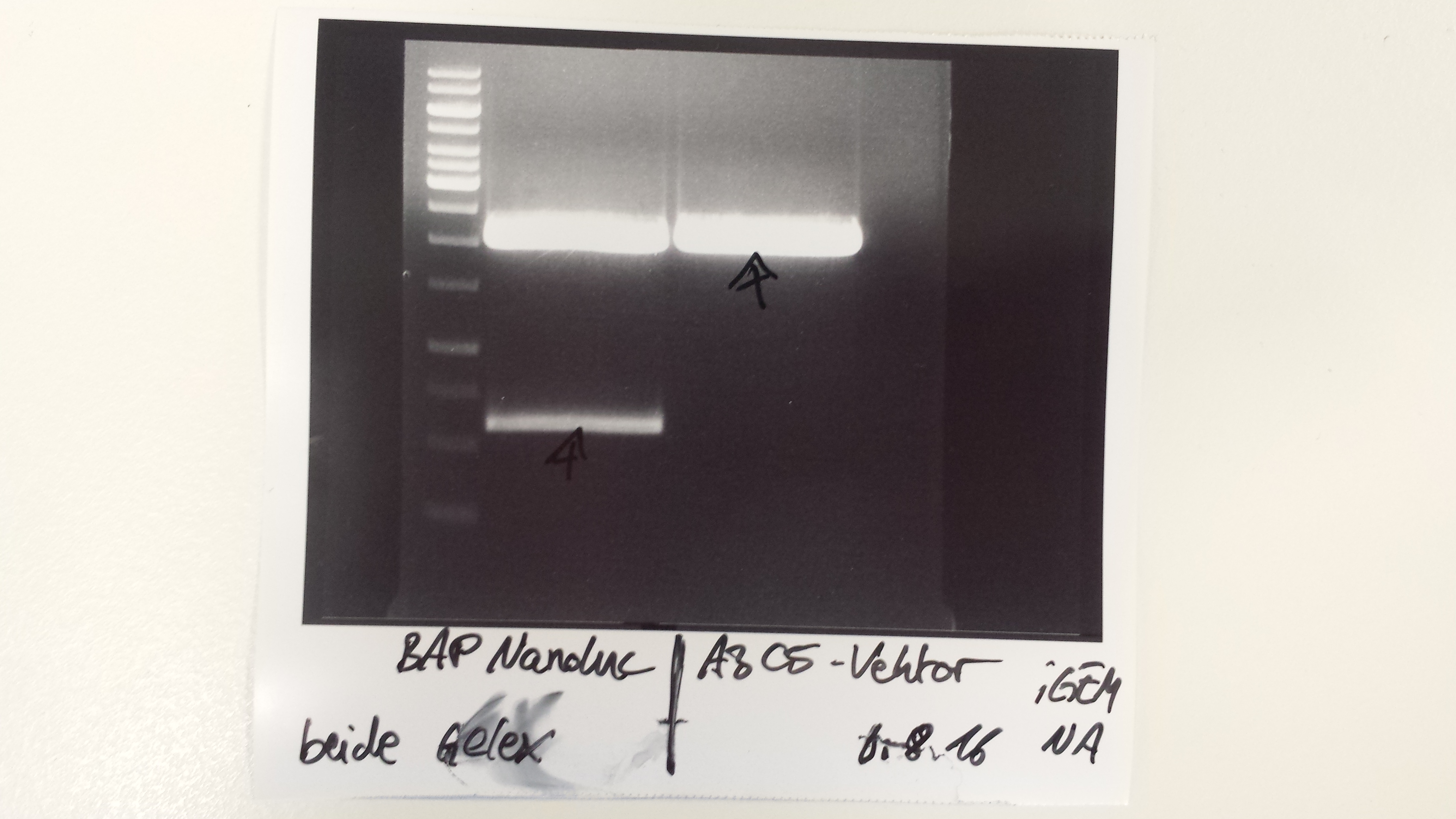
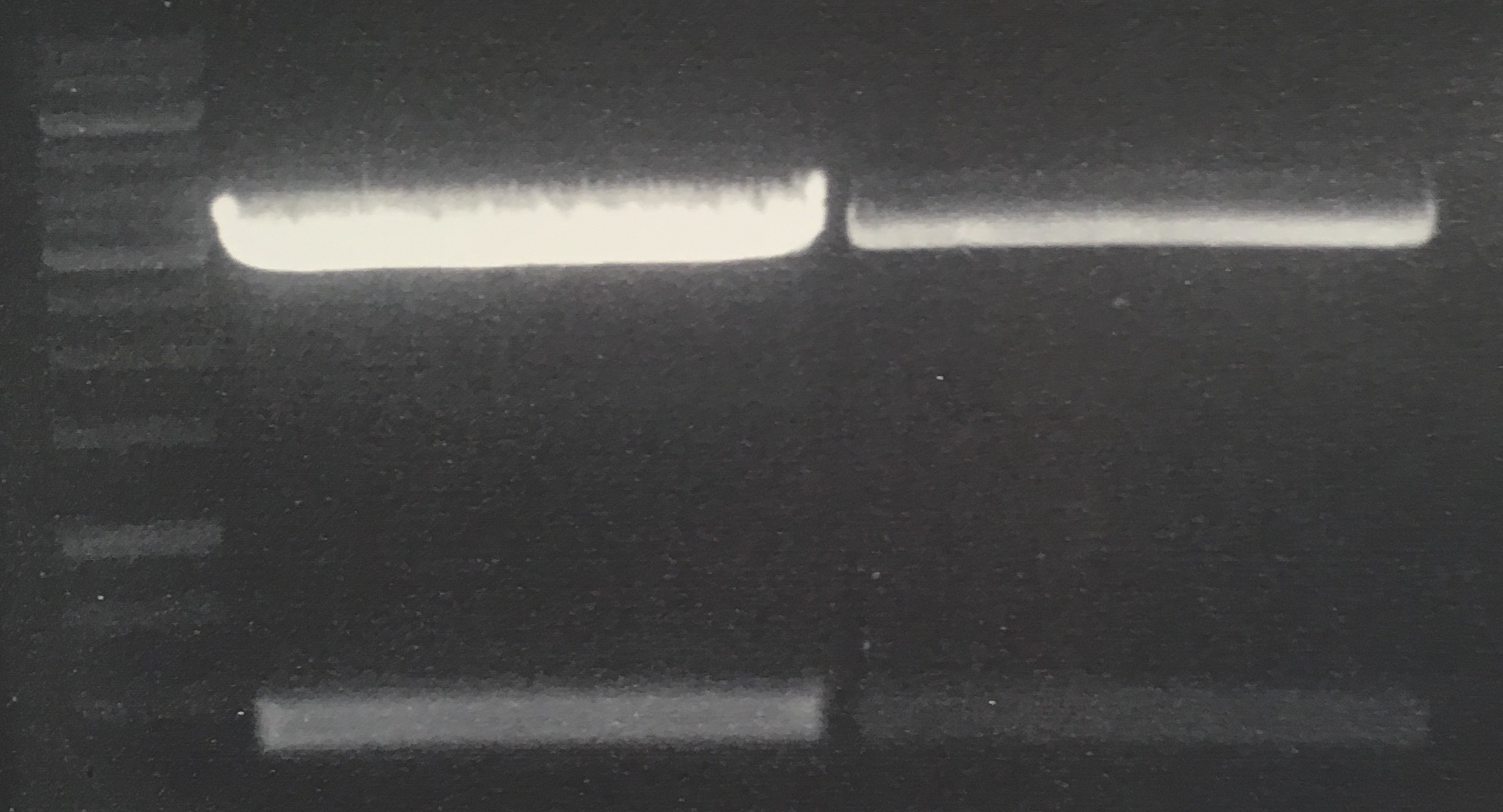
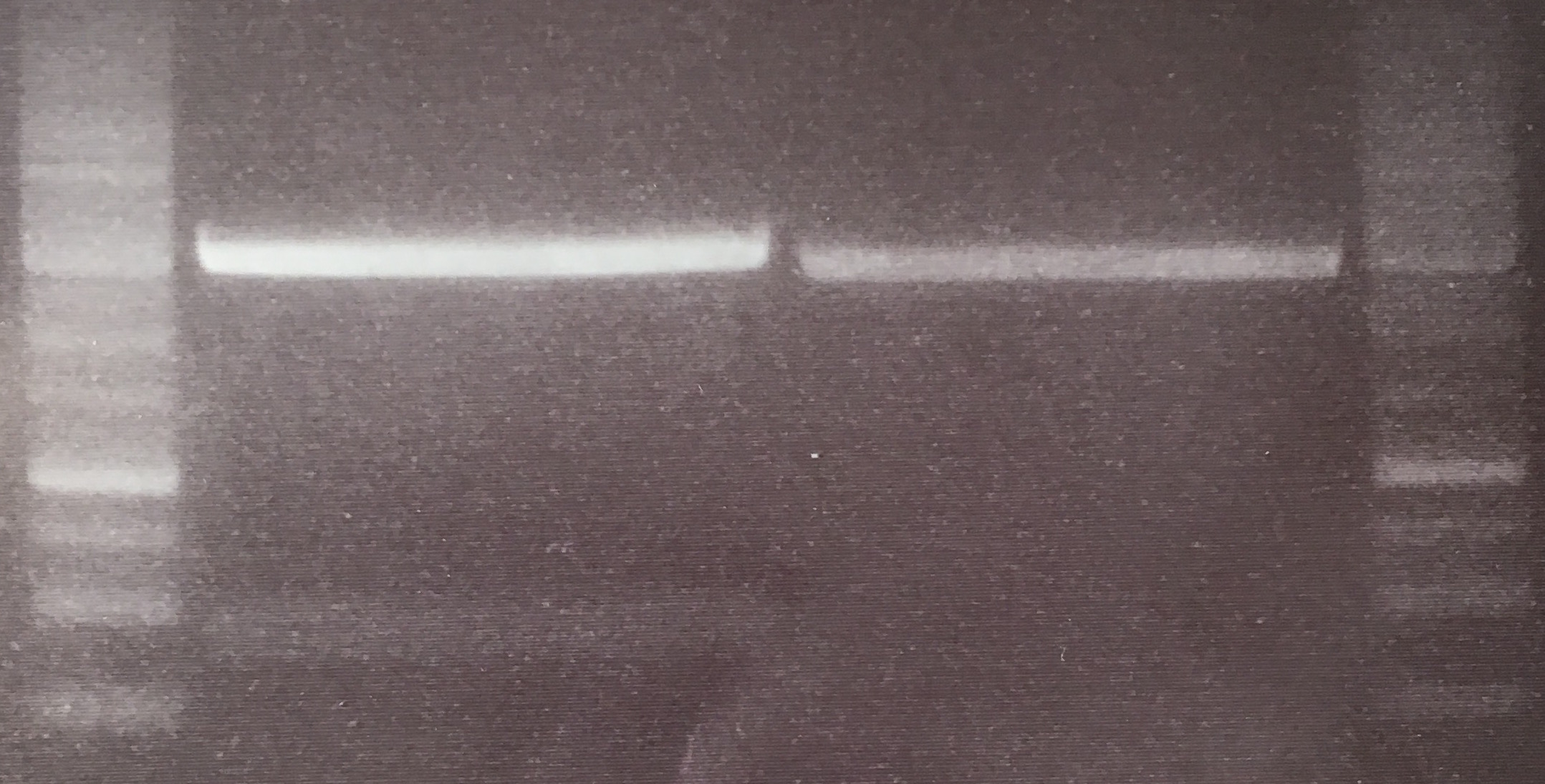
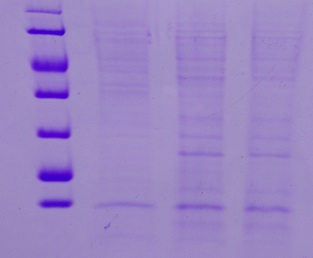
(→Transformation of E. coli XL1 blue with EreA and EreB (pDEST14)) |
(→Monday, Aug 22nd) |
||
| Line 1: | Line 1: | ||
| − | {{LMU-TUM_Munich}} | + | {{LMU-TUM_Munich|navClass=labjournal}} |
{{LMU-TUM_Munich/16style}} | {{LMU-TUM_Munich/16style}} | ||
{{Team:TU-Munich/ExCol}} | {{Team:TU-Munich/ExCol}} | ||
{{Team:TU-Munich/LabHeader}} | {{Team:TU-Munich/LabHeader}} | ||
| − | + | <html> | |
| − | + | </div> | |
| + | <div class="col-sm-8 col-sm-offset-3"> | ||
| + | </html> | ||
<div id="wikicontent-container"> | <div id="wikicontent-container"> | ||
<div id="wikicontent"> | <div id="wikicontent"> | ||
| − | == Labjournal == | + | ==Labjournal== |
| + | |||
<html> | <html> | ||
| Line 13: | Line 16: | ||
<b>Display:</b><br> | <b>Display:</b><br> | ||
<input class="labcheckbox" type="checkbox" name="category" value="general" id="ui-test7" /><b style="color: rgb(115, 208, 255);">General</b><br> | <input class="labcheckbox" type="checkbox" name="category" value="general" id="ui-test7" /><b style="color: rgb(115, 208, 255);">General</b><br> | ||
| − | <input class="labcheckbox" type="checkbox" name="category" value=" | + | <input class="labcheckbox" type="checkbox" name="category" value="streptavidin" id="ui-test1" /><b style="color: rgb(166, 126, 166);">Streptavidin</b><br> |
| − | <input class="labcheckbox" type="checkbox" name="category" value=" | + | <input class="labcheckbox" type="checkbox" name="category" value="linkers" id="ui-test2" /><b style="color: rgb(116, 183, 112);">Linkers</b><br> |
| − | <input class="labcheckbox" type="checkbox" name="category" value=" | + | <input class="labcheckbox" type="checkbox" name="category" value="receptor" id="ui-test3" /><b style="color: rgb(255, 122, 97);">Receptor</b><br> |
| − | <input class="labcheckbox" type="checkbox" name="category" value=" | + | <input class="labcheckbox" type="checkbox" name="category" value="optogenetics" id="ui-test5" /><b style="color: rgb(0, 32, 96);">Optogenetics</b><br> |
| + | <input class="labcheckbox" type="checkbox" name="category" value="polymerization" id="ui-test10" /><b style="color: rgb(160, 0, 39);">Polymerization</b><br> | ||
| + | <input class="labcheckbox" type="checkbox" name="category" value="hypoxia" id="ui-test11" /><b style="color: rgb(219, 48, 130);">Hypoxia</b><br> | ||
| + | <input class="labcheckbox" type="checkbox" name="category" value="kill-switch" id="ui-test12" /><b style="color: rgb(255, 255, 0);">Kill-Switch</b><br> | ||
<a href="#" id="ExAll">Expand All ...</a><br> | <a href="#" id="ExAll">Expand All ...</a><br> | ||
| Line 22: | Line 28: | ||
<b>Jump to:</b><br> | <b>Jump to:</b><br> | ||
| − | <a href="#Week_1">Week 1</a> | + | <a href="#Week_0">Week 0</a> sample<br> |
| − | <a href="#Week_2">Week 2</a> | + | <a href="#Week_1">Week 1</a> May 16–22<br> |
| − | <a href="#Week_3">Week 3</a> | + | <a href="#Week_2">Week 2</a> May 23–29<br> |
| − | <a href="#Week_4">Week 4</a> | + | <a href="#Week_3">Week 3</a> May 30–June 05<br> |
| − | <a href="#Week_5">Week 5</a> | + | <a href="#Week_4">Week 4</a> June 06–12<br> |
| − | <a href="#Week_6">Week 6</a> | + | <a href="#Week_5">Week 5</a> June 13–19<br> |
| − | <a href="#Week_7">Week 7</a> 13 | + | <a href="#Week_6">Week 6</a> June 20–26<br> |
| + | <a href="#Week_7">Week 7</a> June 27–July 03<br> | ||
| + | <a href="#Week_8">Week 8</a> July 04–10<br> | ||
| + | <a href="#Week_9">Week 9</a> July 11–17<br> | ||
| + | <a href="#Week_10">Week 10</a> July 18–24<br> | ||
| + | <a href="#Week_11">Week 11</a> July 25–31<br> | ||
| + | <a href="#Week_12">Week 12</a> Aug 01–07<br> | ||
| + | <a href="#Week_13">Week 13</a> Aug 08–14<br> | ||
| + | <a href="#Week_14">Week 14</a> Aug 15–21<br> | ||
| + | <a href="#Week_15">Week 15</a> Aug 22–28<br> | ||
| + | <a href="#Week_16">Week 16</a> Aug 29–Sep 04<br> | ||
| + | <a href="#Week_17">Week 17</a> Sep 05–11<br> | ||
| Line 35: | Line 52: | ||
<div id="ladder" class="ui-widget ui-widget-content ui-corner-right"> | <div id="ladder" class="ui-widget ui-widget-content ui-corner-right"> | ||
| − | < | + | <br><b>1 kbp GeneRuler:</b><br><br> |
<img src="https://static.igem.org/mediawiki/2012/e/e3/TUM12_1000bp.jpg"></img><br> | <img src="https://static.igem.org/mediawiki/2012/e/e3/TUM12_1000bp.jpg"></img><br> | ||
| − | < | + | <br><b>100 bp GeneRuler:</b><br><br> |
<img src="https://static.igem.org/mediawiki/2012/d/da/TUM12_100bp.jpg"></img><br> | <img src="https://static.igem.org/mediawiki/2012/d/da/TUM12_100bp.jpg"></img><br> | ||
| − | < | + | <br><b>PageRuler Plus:</b><br><br> |
<img src="https://static.igem.org/mediawiki/2012/8/8c/TUM12_250kDa.jpg"></img><br> | <img src="https://static.igem.org/mediawiki/2012/8/8c/TUM12_250kDa.jpg"></img><br> | ||
</div> | </div> | ||
| Line 47: | Line 64: | ||
</div> | </div> | ||
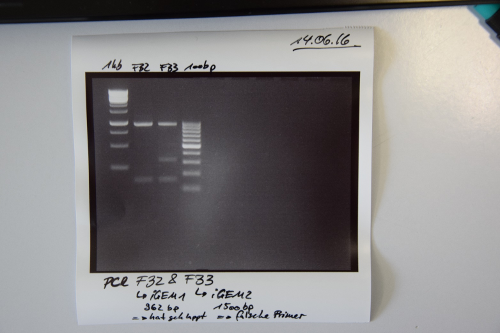
| − | = | + | =Samples= |
| − | <div class="week" id=" | + | <div class="week" id="Week_0"> |
| − | + | ||
| − | <div class=" | + | <div class="linkers"> |
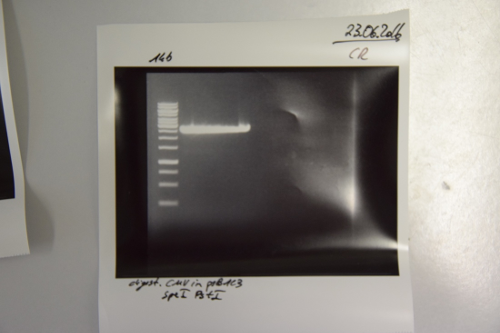
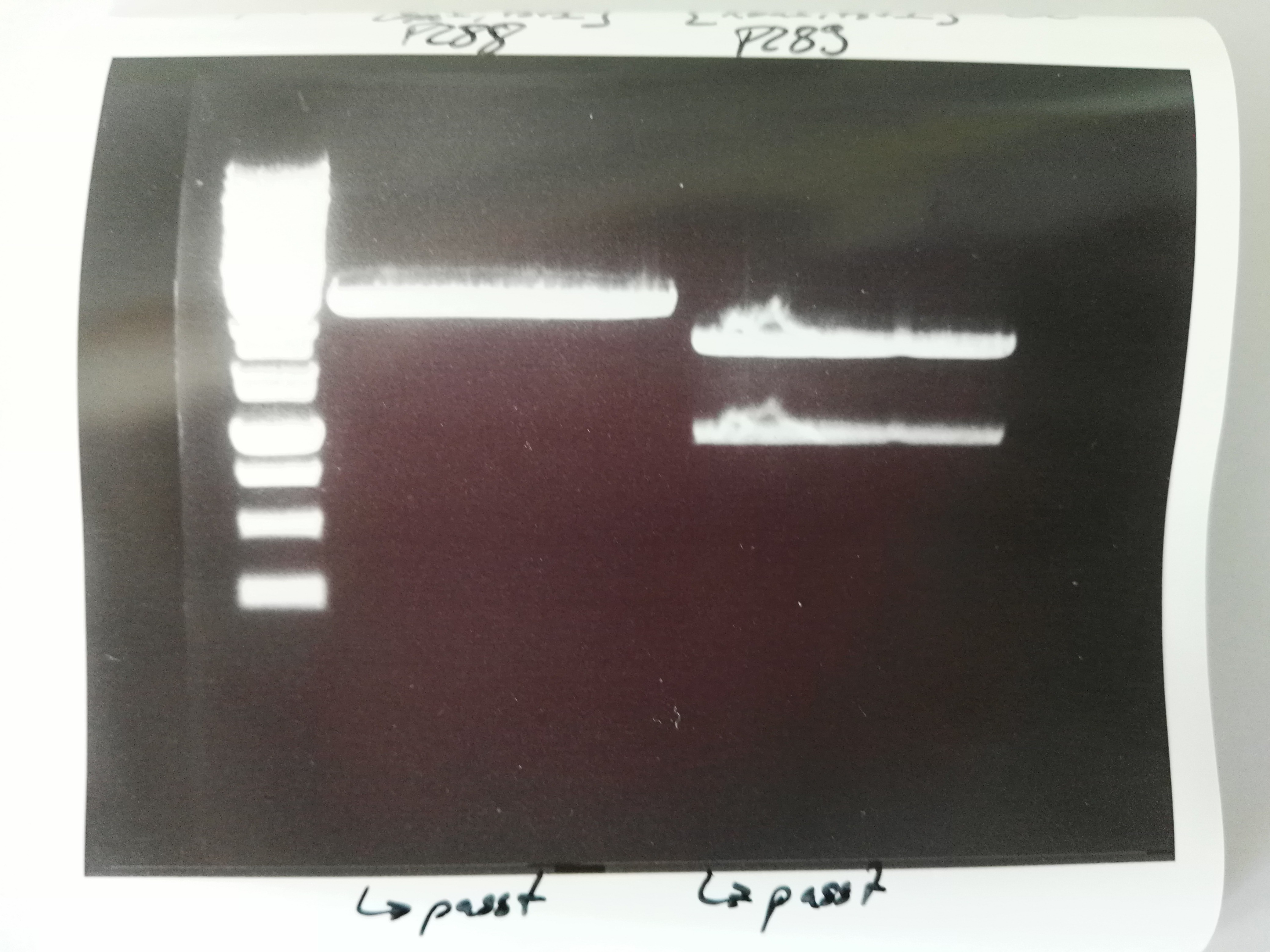
| − | === Transformation of ''E. coli'' XL1 blue with Phytochrome B (2 | + | === Transformation of ''E. coli'' XL1 blue with Phytochrome B (2–908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === |
'''Investigator: Jeff, Rosario''' | '''Investigator: Jeff, Rosario''' | ||
| − | '''Aim of the experiment:''' Transformation of Phytochrome B for protein fusion | + | '''Aim of the experiment:''' Transformation of Phytochrome B for protein fusion |
'''Procedure:''' | '''Procedure:''' | ||
| − | * | + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice |
| − | * 2 µl of DNA was added to 100 µl of competent cells and gently mixed | + | * 2 µl of DNA was added to 100 µl of competent cells and gently mixed |
* 30 min incubation on ice | * 30 min incubation on ice | ||
| − | * 5 min | + | * 5 min heat shock at 37 °C |
| − | * | + | * Added 1 ml LB medium to each tube |
| − | * Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker | + | * Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker |
| − | * 100 µl of the cell suspension was plated on one chloramphenicol plate | + | * 100 µl of the cell suspension was plated on one chloramphenicol plate |
| − | * The rest were centrifuged for 1 min at | + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded |
* The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate. | * The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate. | ||
| Line 105: | Line 121: | ||
</div> | </div> | ||
| − | + | <div class="optogenetics"> | |
| − | + | ||
| − | <div class=" | + | |
=== Picking of of ''E. coli'' XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | === Picking of of ''E. coli'' XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| Line 234: | Line 248: | ||
</div> | </div> | ||
| − | + | <div class="receptor"> | |
| − | + | ||
| − | <div class=" | + | |
=== Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | === Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3) === | ||
| Line 250: | Line 262: | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="linkers"> |
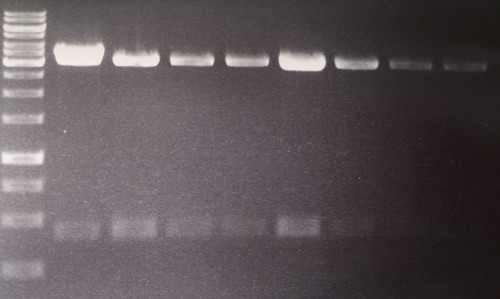
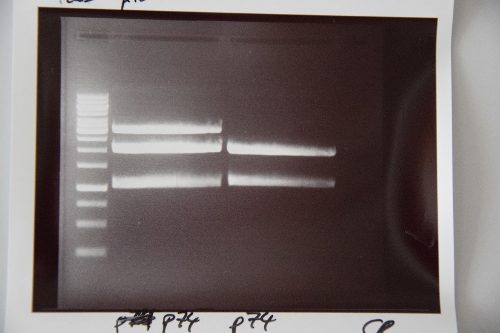
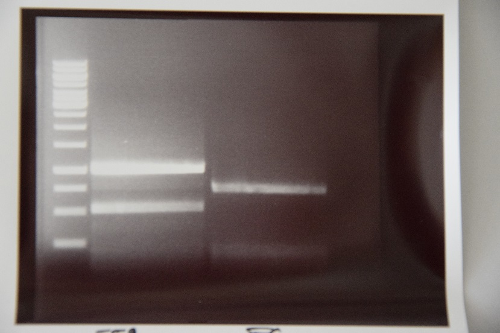
=== Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10 === | === Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10 === | ||
| Line 381: | Line 393: | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="linkers"> |
| − | === Transformation of ''E. coli'' XL1 blue with | + | === Transformation of ''E. coli'' XL1 blue with === |
| − | '''Investigator: | + | '''Investigator: ''' |
'''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| Line 393: | Line 405: | ||
* CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| − | * | + | * µl of DNA was added to 100 µl of competent cells and gently mixed. |
* 30 min incubation on ice | * 30 min incubation on ice | ||
| Line 399: | Line 411: | ||
* 5 min. heat shock at 37 °C | * 5 min. heat shock at 37 °C | ||
| − | * Adding of | + | * Adding of µl LB-medium to each tube. |
* The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| Line 410: | Line 422: | ||
<!--- PLEASE DO NOT TOUCH !!!! --> | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| − | =Week | + | =Week 1, May 16th - May 22nd= |
| − | <div class="week" id=" | + | <div class="week" id="Week_1"> |
| − | + | ||
| − | <div class=" | + | =='''Monday, May 16th'''== |
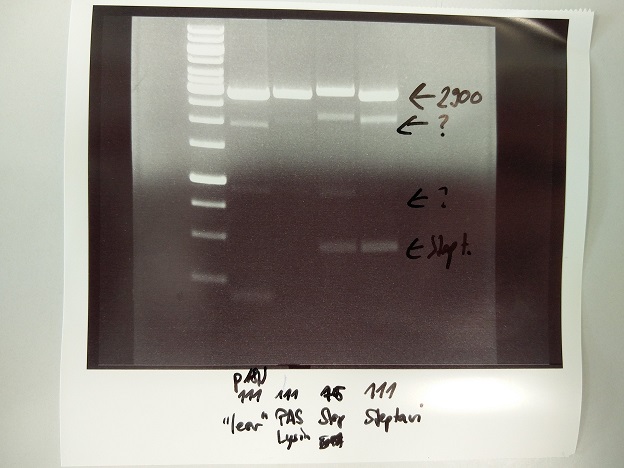
| + | <div class="streptavidin"> | ||
| + | === Streptavidin Plasmid Check === | ||
| + | '''Investigator: JB, LK, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verify cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep following manufacturer's protocol (QIAprep MiniPrep, Qiagen) (4 clones each of pSA1, pSAm1 in pASK75) | ||
| + | * Analytic digestion with 0.25 ''µ''L XbaI, 0.25 ''µ''L HindIII (HF), 1 ''µ''L SmartCut Buffer, 5 ''µ''L plasmid DNA, 3.5 ''µ''l H<sub>2</sub>O | ||
| + | * 5 ''µ''L analyzed in gel electrophoresis (1% agarose) | ||
| + | |||
| + | '''Results:''' Successful cloning verified. Stored at –20 °C | ||
| + | |||
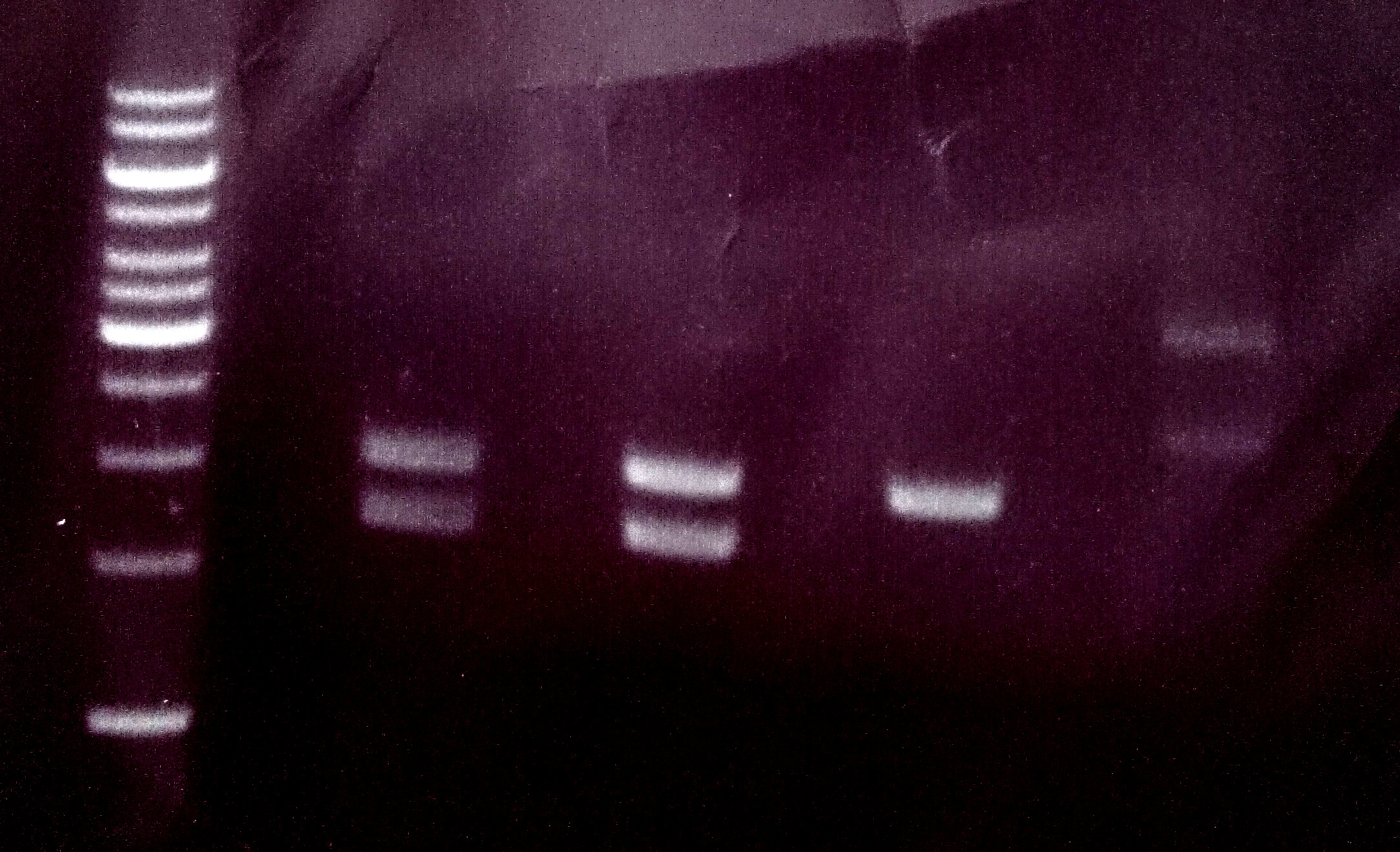
| + | * Lane 1: 5 ''µ''L Thermo Fisher, 1 kb ladder | ||
| + | |||
| + | * Lanes 2–9: 5 ''µ''L digestions of P6–P13, band of SA (mut1) at about 300 bp, band of digested plasmid at about 3,000 bp | ||
| + | |||
| + | |||
| + | [[File:Muc16_P6_to_P13_SA_mut1.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Streptavidin Expression: Transform BL21 === | ||
| + | |||
| + | '''Investigator: JB, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Express pSA1 and pSAm1 in ''E. coli'' BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Transformation into competent ''E. coli'' BL21 according to protocol of P6 and P10 | ||
| + | |||
| + | '''Results:''' Plates (LB Amp) in incubator (37 °C) for further processing | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, May 17th'''== | ||
| + | <div class="general"> | ||
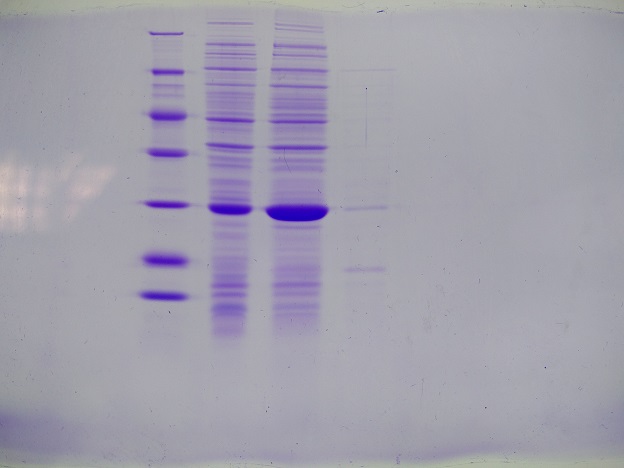
| + | === SDS Gel Analysis === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analyze collagen 1/2, eGFP, fraction 30 of egg precipitation ''via'' SDS gel | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Mixed 80 ''µ''L samples with 20 ''µ''L SDS buffer | ||
| + | |||
| + | * Heated at 95 °C for 10 min | ||
| + | |||
| + | * 1 d staining, 1 d unstaining | ||
| + | |||
| + | '''Results:''' Successful cloning verified. Stored at –20 °C | ||
| + | |||
| + | * Lane 1: 8 ''µ''L marker (Thermo Fisher #26610) | ||
| + | |||
| + | * Lane 2: Fraction 30 (IEC), 3 ''µ''L, band at 35 kDa, avidin expected at 16 kDa | ||
| + | |||
| + | * Lane 3: eGFP, 12 ''µ''L, band at 27 kDa, eGFP expected at 27 kDa, many impurities | ||
| + | |||
| + | * Lane 4: Collagen 1, 12 ''µ''L, no sharp band | ||
| + | |||
| + | * Lane 5: Collagen 1, 12 ''µ''L, no sharp band | ||
| + | |||
| + | |||
| + | [[File:Muc16_SDS_gel_analysis_fraction_30_of_egg-precipitation.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
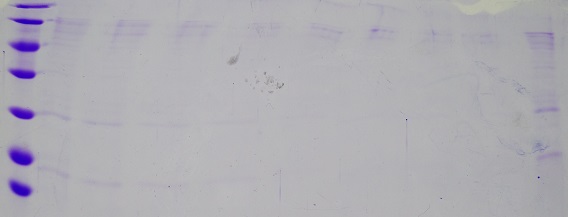
| + | === MiniPreps pSb1C3–AviTag, –A3C5 and pASK75–(SA1), –(SAm1) === | ||
| + | |||
| + | '''Investigator: CR, CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verify cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed following manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | |||
| + | * Analytic digestion with 0.25 ''µ''L XbaI, 0.25 ''µ''L HindIII (HF) for pASK plasmids and 0.25 ''µ''L NgomIV, 0.25 ''µ''L AgeI (HF) for pSb1C3 plasmids, 1 ''µ''L SmartCut Buffer, 5 ''µ''L plasmid DNA, 3.5 ''µ''L H<sub>2</sub>O | ||
| + | |||
| + | * 5 ''µ''L on 1% agarose gel electrophoresis | ||
| + | |||
| + | '''Results:''' Successful cloning verified for pASK plasmids. Repeat pSb1C3 plasmids. Stored at –20 °C | ||
| + | |||
| + | * Lane 1: 5 ''µ''L Thermo Fisher, 1 kb Ladder | ||
| + | |||
| + | * Lane 2: 5 ''µ''L digestion of pSb1C3–AviTag | ||
| + | |||
| + | * Lane 3: 5 ''µ''L digestion of pSb1C3–A3C5 | ||
| + | |||
| + | * Lane 5: 5 ''µ''L digestion of pASK75(SA1), EB elution | ||
| + | |||
| + | * Lane 6: 5 ''µ''L digestion of pASK75(SAmut1), EB elution | ||
| + | |||
| + | * Lane 7: 5 ''µ''L digestion of pASK75(SAmut1), H<sub>2</sub>O elution | ||
| + | |||
| + | |||
| + | [[File:Muc16_verification_pASK.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inoculation of Pre-Culture with BL21 (pASK75 (SA1)) in LB Medium === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Pre-culture for streptavidin expression in TB medium | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Added 50 ''µ''L ampicillin to 50 mL LB medium | ||
| + | |||
| + | * Picked colonies from BL21 (pASK75 (SA1)) | ||
| + | |||
| + | * Inoculated LB medium | ||
| + | |||
| + | * Incubated at 30 °C overnight | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, May 18th'''== | ||
| + | <div class="general"> | ||
| + | === Repeat Analytical Gel of pSb1C3–AviTag, –A3C5 === | ||
| + | |||
| + | '''Investigator: CG, CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verify cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Analytic digestion with 0.25 ''µ''L NgoMIV, 0.25 ''µ''L AgeI (HF) for pSb1C3 plasmids, 1 ''µ''L SmartCut Buffer, 8.5 ''µ''L plasmid DNA | ||
| + | * 10 ''µ''L analyzed in gel electrophoresis (2% agarose) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16 Repetition of analytical gel of pSb1C3-AviTag,-A3C5.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inoculation of BL21 (pASK75 (SA1)) Culture in 2 L TB Medium and Induction of Streptavidin Production by Addition of Tetracycline === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Produce streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Ampicillin (2 mL) was added to the medium (1:1000) | ||
| + | * The pre-culture (50 mL) was poured into the medium | ||
| + | * Culture incubated at 37 °C and 140 rpm until OD<sub>550</sub> reached 0.5 | ||
| + | * To induce streptavidin expression, anhydro-tetracycline (200 ''µ''L) was added to the culture (1:10,000) | ||
| + | * The culture was incubated at 37 °C and 140 rpm for 4 hours | ||
| + | |||
| + | '''Results:''' Streptavidin expression by BL21 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Expression and Harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB Medium === | ||
| + | |||
| + | '''Investigator: CR, JB, JH ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor) | ||
| + | * The supernatant was discarded, and the pellet was transferred into a beaker of sufficient size and resuspended in Tris Buffer B cooled to 4 °C (50 mM Tris/HCl pH = 8.0, 1 mM EDTA) | ||
| + | * The solution was homogenized in the PANDA (ask supervisor) | ||
| + | * The resulting lysate was transferred into centrifuge tubes and spun down (4 °C, 18,000 rpm, 10 min, XX34-rotor). The supernatant was discarded, and the pellet was resuspended in 6 M Gua-HCl (pH = 1.5) at 4 °C overnight. | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Dialysis of eGFP === | ||
| + | |||
| + | '''Investigator: NA, JH, CR ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purify eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * eGFP was thawed on ice | ||
| + | * eGFP was then poured into a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in ice-cold Tris/HCl 20 mM pH 8.0 | ||
| + | * Dialyzed at 4 °C overnight (XX34 rotor). The supernatant was discarded, and the pellet was | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === MiniPrep of quickchanged pNGAL146-A2 EspP === | ||
| + | |||
| + | '''Investigator: NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extraction of pNGAL146-A2 plasmid from XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
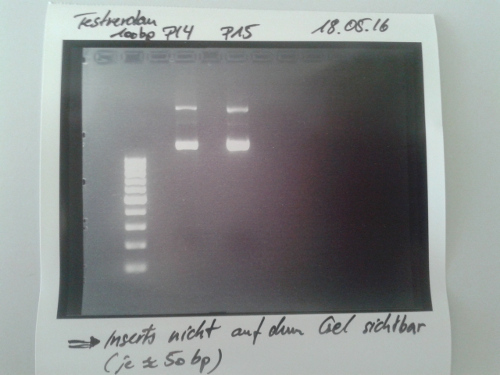
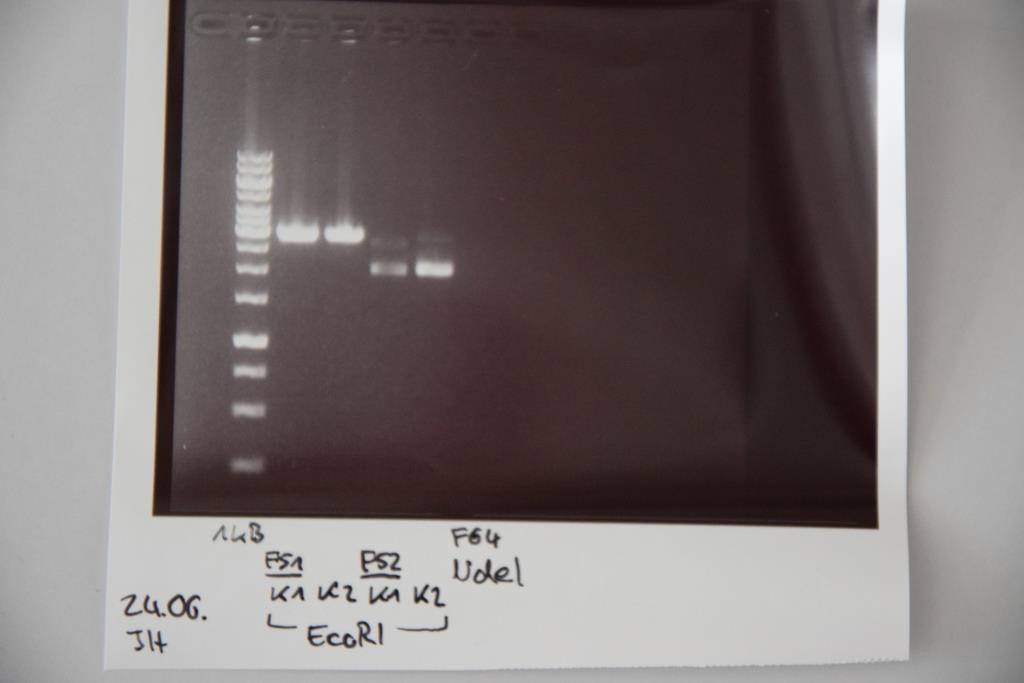
| + | === Sequencing of P14 (Avi-Tag), P15 (A3C5) & P19 (Quick change (QC) EspP in pNGAL-A2) === | ||
| + | |||
| + | '''Investigator: CR, NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P14 (Avi-Tag), P15 (A3C5) & P19 (Quick change (QC) EspP in pNGAL-A2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | * The different plasmids we prepared received the following barcodes: | ||
| + | * P14 : FR11326653 | ||
| + | * P15 : FR11326655 | ||
| + | * P19 (K4): FR11326654 | ||
| + | * P16 (K1): FR11326652 | ||
| + | * P17 (K2): FR11326651 | ||
| + | * P18 (K3): FR11326650 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
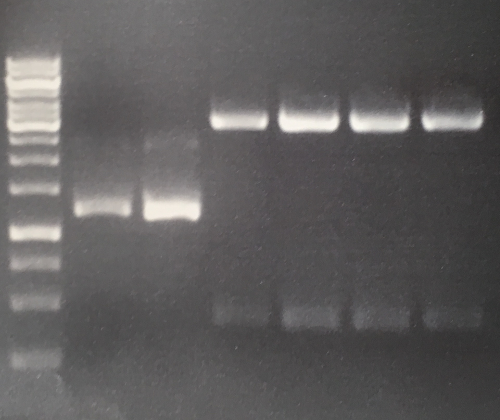
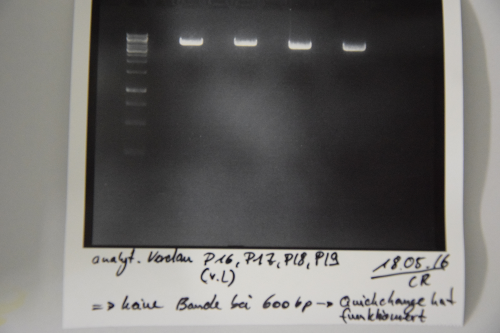
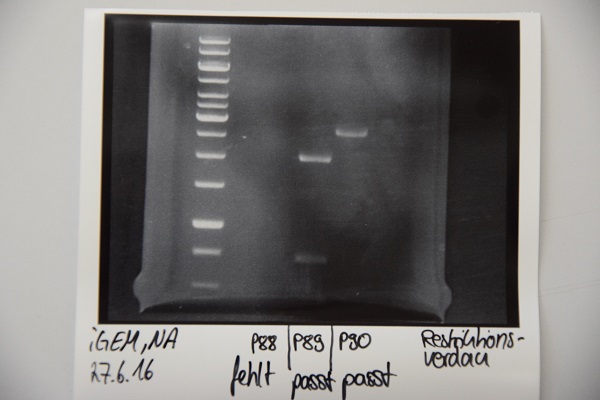
| + | === Digestion of P16 , P17, P18 & P19 (all QC EspP in pNGAL-A2) with AgeI & HindIII + analytical gel === | ||
| + | |||
| + | '''Investigator: NA, JH, CR ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of success of quickchange | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
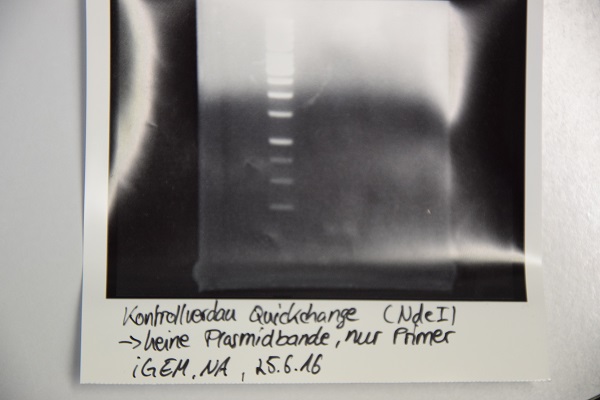
| + | * analytic digestion with: 0,25 µl HindIII (HF), 0,25 µl AgeI (HF), 1 µl SmartCut Buffer, 500 ng plasmid-DNA, fill up with ddH2O (Vtotal= 10µL) | ||
| + | * 10 µl on 2% agarose gel for electrophoresis | ||
| + | |||
| + | '''Results:''' No signal at 600 bp --> quickchange seems to be successful (waiting for sequencing) | ||
| + | |||
| + | |||
| + | [[File:Muc16_Verification_of_success_of_quickchange.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, May 19th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Re-Sequencing of P19=== | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Re-Sequencing of P19 | ||
| + | |||
| + | '''Procedure:'''Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | |||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | *P19 (K4): FR11326649 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Cloning of A3C5 and Avi-Tag into pSB1C3=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Re-Trafo of pSB1C3 RFP for later on: digestion, dephosphorylation and cloning | ||
| + | |||
| + | '''Procedure:''' transformation according to protocol of P4 E. Coli XL1 | ||
| + | |||
| + | '''Result:''' plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | ===Streptavidin refolding=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Refolding of denaturated Streptavidin | ||
| + | |||
| + | '''Procedure:''' After the pellet had almost completely dissolved in 6M GdmCl, the solution was spun down (4°C, 20 mins, 18,000 rpm). The supernatant was transferred carefully into a falcon tube and the pellet was cast away. Via a hydraulic pump (flow rate: 2x10 ml/min) the lysate was transferred Into 5L PBS 1x. Afterwards the pump was cleaned with technical isopropanol and ELGA water. The solution was stirred overnight at 4°C for refolding. | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | ===Biotinylation of BSA=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Biotinylation of BSA | ||
| + | |||
| + | '''Procedure:''' A 100 µM (=6.8 mg/ml) solution of BSA (Albumin fraction V, pH=7, in the fridge in the central lab) was created (V=10 ml). 220 µl of a 100 mM Biotin stock were added. The mixture was stored overnight Iin the fridge (4°C). | ||
| + | |||
| + | '''Result:''' Hopefully biotinylated BSA mixture in the fridge (4°C). | ||
| + | </div> | ||
| + | |||
| + | =='''Friday, May 20th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Qualitative analysis of streptavidin expression === | ||
| + | |||
| + | '''Investigator: CG ''' | ||
| + | |||
| + | '''Aim of the experiment:''' SDS gel analysis of recombinant strepatividin expression | ||
| + | |||
| + | '''Procedure:''' mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining | ||
| + | |||
| + | '''Result:''' | ||
| + | |||
| + | * 1. Lane: 8 µl Marker (Thermo Fisher #26610) | ||
| + | * 2. Lane: 12 µl culture aliquot, before induction | ||
| + | * 3. Lane: 12 µl culture aliquot, after induction | ||
| + | * 4. Lane: 3 µl culture aliquot of the lysed pelet, Strepatvidin expected at about 16 kDa | ||
| + | * 5. Lane: 3 µl culture aliquot of the supernatend after lysis, no Strepatvidin expected at about 16 kDa | ||
| + | * 6. Lane: 1.5 µl culture aliquot of the lysed pelet, Strepatvidin expected at about 16 kDa | ||
| + | * 7. Lane: 1.5 µl culture aliquot of the supernatend after lysis, no Streptavidin expected at about 16 kDa | ||
| + | |||
| + | [[File:Muc16_SDS_gel_analysis_of_recombinant_strepatividin_expression.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | <div class="receptor"> | ||
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Inoculation of pre-culture with E. coli XL1 (pSb1C3 -RFP) in LB-Chloramphenicol-medium | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Picking of colonies for E. coli XL1 (pSb1C3 -RFP) | ||
| + | Inoculate in 5 ml LB-Chloramphenicol-medium | ||
| + | Incubate at 37°C over night at 200 rpm | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of Biobricks in XL1-blue === | ||
| + | |||
| + | '''Investigator: NA, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | -10 µl dd H2O in well of interest (standard distribution kit) | ||
| + | -1 µl Plasmid (out of well) to cells | ||
| + | -Transformation according to the SOP | ||
| + | Used bricks: | ||
| + | K577893, B0015, R0040, B0032, I14033, K747096 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Ammonium sulfate precipitation of streptavidin === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Reduction of the protein solution volume and precipitation of streptavidin | ||
| + | |||
| + | '''Procedure:''' The 5 L protein solution was spun down (20 mins, 5,000 rpm) and the supernatant transferred into a beaker. In order to lower the volume of the solution for ammonium sulfate precipitation, the solution was first filtered via a membrane crossflow pump (membrane: Sartocon 0.45 µm, thick membrane). | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Dialysis of biotinylated BSA === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of biotinylated BSA | ||
| + | |||
| + | '''Procedure:''' dialysis against Tris/HCl 20 mM, pH 8, 10 mM NaCl over night; | ||
| + | cut off : 14 kDa | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 2 (May 23rd - May 29th)= | ||
| + | <div class="week" id="Week_2"> | ||
| + | |||
| + | =='''Monday, May 23th'''== | ||
| + | |||
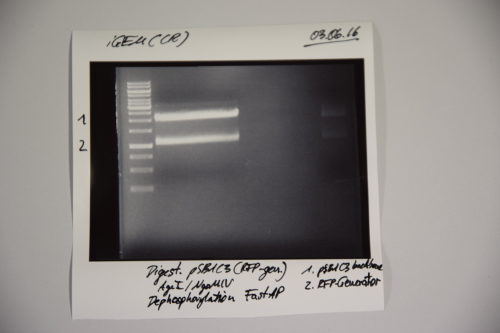
| + | <div class="receptor"> | ||
| + | ===Cloning of A3C5 and Avi-Tag into pSB1C3=== | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Cloning of A3C5 and Avi-Tag into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * pSB1C3 was digested with AgeI and NgOMIV (10 µL plasmid + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 14 µL ddH2O; incubation for 1h, 37°C) | ||
| + | * FastAP was added and the reaction was incubated for another 2h at 37°C | ||
| + | * The digestion was purified by gelelctrophoresis (1%, 75V, 1h) | ||
| + | * Plasmid backbone was cut from the gel | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Cloning_of_A3C5_and_Avi-Tag_into_pSB1C3.png|500px]] | ||
| + | |||
| + | Signal 1: linearized pSB1C3 (successfull digestion) | ||
| + | |||
| + | Signal 2: supercoiled pSB1C3 (digestion not successfull) | ||
| + | |||
| + | Signal 3: RFP-generator | ||
| + | |||
| + | --> next time gel should run longer (just if you need the back bone) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Filtration of Streptavidin and precipitation with Ammonium-sulfate === | ||
| + | |||
| + | '''Investigator: MP, CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Concentration of Streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | The solution was filtered via a membrane crossflow pump (membrane: Sartocon 0.45 µm, thick membrane). | ||
| + | The final volum was 0.5L | ||
| + | The solution was precipitated by addition of Ammonium-sulfate (2 steps: 1. 40% Ammonium-sulfate; 2. 70% Ammonium-sulfate) | ||
| + | After the first addition of ammonium-sulfate the solution was stirred (1h, 4°C) | ||
| + | Afterwards the solution was centrifuged (10.000 rpm; 30 min) and the supernatant was used for the second precipitation step | ||
| + | The solution was stirred again (over night, 4°C) | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Inoculation of pre-culture with BioBricks in pSB1C3 in LB-Cam === | ||
| + | |||
| + | '''Investigator: JH, JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Add 50 µL chloramphenicol in 50 mL LB-medium | ||
| + | Picking colonies from K577893; B0015; B0032 (in pSB1C3) * | ||
| + | Inoculate 4 mL medium (CAM) | ||
| + | Incubate at 37°C over night | ||
| + | *Following cultures didn´t grow and were therefore not inoculated: | ||
| + | R0040; K747096; I14033 | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Retransformation of Biobricks in XL1-blue=== | ||
| + | |||
| + | '''Investigator: JH, JL, EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | 10 µl dd H2O in well of interest (standard distribution kit) | ||
| + | 1 µl Plasmid (out of well) to cells | ||
| + | Transformation according to the SOP | ||
| + | Used bricks: | ||
| + | K577893, B0015, R0040, B0032, I14033, K747096 | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, May 24th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of pre-culture with BioBricks in pSB1C3 in LB-Cam === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | '''Procedure: ''' | ||
| + | Add 50 µL chloramphenicol in 50 mL LB-medium | ||
| + | Picking colonies from K577893; B0015; B0032 (in pSB1C3) * | ||
| + | Inoculate 4 mL medium (CAM) | ||
| + | Incubate at 37°C over night | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Gelextraction of Signal 1 from pSB1C3-digestion | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen) | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Ammonium sulfate precipitation === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Precipitation the 70% saturated ammonium sulfate solution | ||
| + | |||
| + | '''Procedure:''' The ammonium sulfate solution, which actually was a suspension, was transferred into centrifuge tubes and spun down (10,000 rpm, 45 mins). As it turns out, unfortunately too much ammonium sulfate was used for the precipitation (wrong table). The precipitate was brought back into solution (20 mM Tris/HCl, pH 8.0) and loaded onto an SDS gel for analysis, along with other samples (flowthrough of the filtration from the day before). | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MiniPrep of BioBricks in pSB1C3 (inoculated on May 23th) === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extraction of BioBricks B0032, B0015 and K577893 in pSB1C3 from XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | concentrations measured (in ng/µL): | ||
| + | (B0032) 360.2; (B0015) 254.9; (K577893) 99.2 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Sequencing of P23 (weak RBS), P24 (double terminator) & P24 (Tet repressed GFP) === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P23, P24 & P25 (x2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | |||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | * P23 (VF2) : FR11326647 | ||
| + | * P24 (VF2) : FR11326646 | ||
| + | * P25 (VF2) : FR11326645 | ||
| + | * P25 (VR) : FR11326644 | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, May 25th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Inoculation of pre-culture with tet-repressed GFP (P25) in LB-Cam === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Add 50 µL chloramphenicol to 50 mL LB-medium | ||
| + | Picking colonies from plate (BL21) | ||
| + | Incubate over night at 37°C | ||
| + | => dismissed, because plasmid sequence was inaccurate | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MiniPrep of K747096, I14033, R0040, B0015, K577893 and B0032 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment: MiniPrep of BioBricks''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | Concentrations were measured (in ng/µL): | ||
| + | |||
| + | * K747096: 286.2 | ||
| + | * I14033: 246.1 | ||
| + | * R0040: 243.5 | ||
| + | * B0015: 244.1 | ||
| + | * K577893: 440.9 | ||
| + | * B0032: 259.7 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
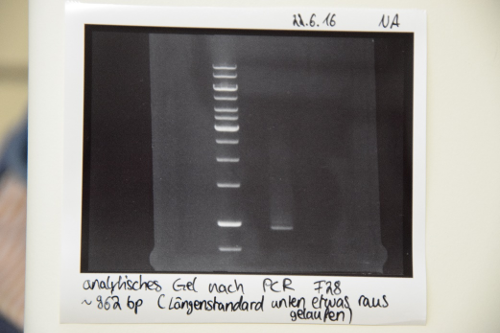
| + | === PCR of P19 (QC EspP) with O3 and O4 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of quickchanged EspP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *reaction was performed after manufacturer's protocol (PCR Using Q5® High-Fidelity DNA Polymerase, NEB) | ||
| + | *274,4 pg Plasmid were used for amplification (1 µL) | ||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial Denaturation||98||30 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |72||30 | ||
| + | |- | ||
| + | |72||60 | ||
| + | |- | ||
| + | |final Extension||72||5 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | * PCR product was purified accoding to manufacturer's protocol (PCR puification kit, Qiagen) | ||
| + | * DNA was eluted 40 µL EB buffer | ||
| + | </div> | ||
| + | <div class="receptor"> | ||
| + | === Digestion of PCR of EspP (P19 with O3 and O4) === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of EspP with AgeI and NgoMIV to ligate it in pSB1C3 later on | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for preparative digestion: | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Volume''' | ||
| + | |'''Reagent''' | ||
| + | |- | ||
| + | |38.5 µL||PCR product P19 with O3 and O4 | ||
| + | |- | ||
| + | |1 µL||AgeI | ||
| + | |- | ||
| + | |1 µL||NgoMIV | ||
| + | |- | ||
| + | |5 µL||CutSmart buffer | ||
| + | |- | ||
| + | |4.5 µL||ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |50 µL||TOTAL | ||
| + | |} | ||
| + | * Incubation for 2 h at 37 °C | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
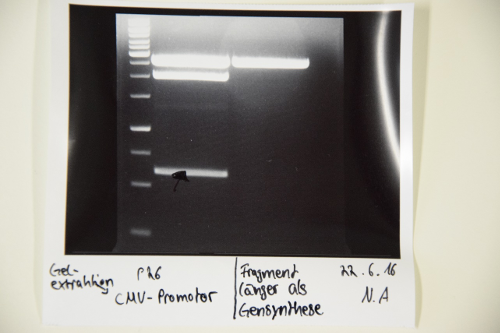
| + | === Sequencing of P26 (CMV promotor), P27 (p cat promotor), P28 (Tet repressible promotor), P30 (Tet repressed GFP) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Sequencing of P26, P27, P28 & P30 (x2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Sequencing batches were prepared after manufacturer's protocol | ||
| + | (15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer) | ||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | P26 (VF2) : FR11326643 | ||
| + | P27 (VF2) : FR11326642 | ||
| + | P28 (VF2) : FR11326641 | ||
| + | P30 (VF2) : FR11326640 | ||
| + | P30 (VR): FR11326639 | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
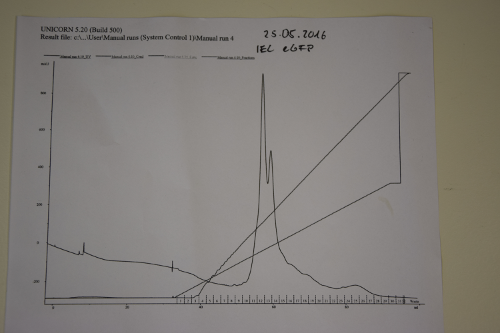
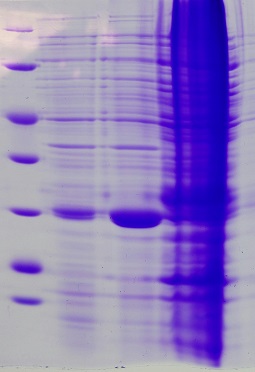
| + | === Ion exchange chromatography of eGFP === | ||
| + | |||
| + | '''Investigator: NA, JB,JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * use Äkta-Purifier (Q-column for eGFP (quarternary ammonium as stationary phase)). Ask Andy before use! | ||
| + | * Pump A: running buffer (20 mM Tris/HCl pH 8); pump B: elution buffer (20 mM Tris/HCl pH 8 and 1,000 mM (1 M) NaCl | ||
| + | * basic pumpwash before start | ||
| + | * injection in loop | ||
| + | * gradient of ion strength on column. | ||
| + | |||
| + | '''Results:''' | ||
| + | Elution of eGFP at ~200 mM NaCl. | ||
| + | |||
| + | [[File:Muc16_elution_of_eGFP.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, May 26th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of cultures with XL-1 F11 and F12 clones === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Inoculation of a 5 ml culture with a clone of the previously transformed F11 (Avi-Tag) F12 (Antibody-binding site). | ||
| + | </div> | ||
| + | <div class="general"> | ||
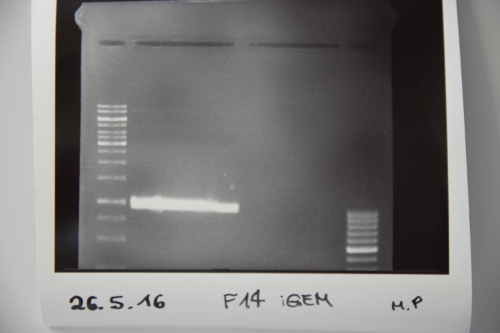
| + | === Cloning of ligation F14 +F13 === | ||
| + | |||
| + | '''Investigator: MP, EF, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' new biobrick EspP in pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | F14 was purified by gelelctrophoresis (1%, 90 V, 40 min) | ||
| + | gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen) | ||
| + | ligation of F14 (EspP) with F13 (dephos. PSB1C3) | ||
| + | transformation according to protocol in competent E.coli XL1 blue | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_F14_gelextracion.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Transformation of Biobrick B0034 in XL1-blue === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µl ddH2O in well of interest (standard distribution kit, plate 4, well 1N) | ||
| + | * 1 µl Plasmid (out of well) to cells | ||
| + | * Transformation according to the SOP (incl. rescue) | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of Biobricks K577895 and K577894 in XL1-blue === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µl ddH2O in well of interest (standard distribution kit, plate 1, wells 14B / 12P) | ||
| + | * 1 µl Plasmid (out of well) to cells | ||
| + | * Transformation according to the SOP (incl. rescue) | ||
| + | </div> | ||
| + | |||
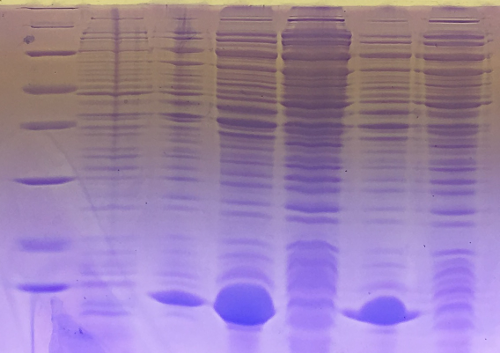
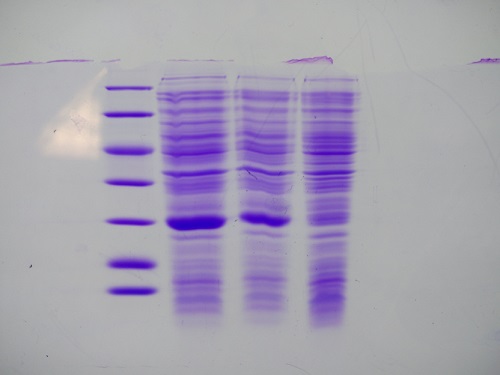
| + | === SDS-PAGE of eGFP-fractions after IEC === | ||
| + | |||
| + | '''Investigator: NA, CG''' | ||
| + | |||
| + | '''Procedure:''' mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining | ||
| + | Concentrations were measured via nanodrop (A280, extinction coefficient: 55000 M−1cm−1) | ||
| + | |||
| + | '''Result:''' concentrations in mg/ml | ||
| + | 1. Lane: 8 µl Marker | ||
| + | 2. Lane: 10 µl of fraction 10; c= 0 | ||
| + | 3. Lane: 10 µl of fraction 11; c= 0,15 | ||
| + | 4. Lane: 10 µl of fraction 12; c= 0,77 | ||
| + | 5. Lane: 10 µl of fraction 13; c= 0,29 | ||
| + | 6. Lane: 10 µl of fraction 14; c= 0,4 | ||
| + | 7. Lane: 10 µl of fraction 15; c= 0,144 | ||
| + | 8. Lane: 10 µl of fraction 16; c= 0,085 | ||
| + | |||
| + | [[File:Muc16_SDS-PAGE_of_eGFP-fractions_after_IEC.png|500px]] | ||
| + | |||
| + | eGFP is clearly detectable at about 27 kDa. The most pure fractions 12 and 13 were pooled for later biotinylation. | ||
| + | </div> | ||
| + | |||
| + | =='''Friday, May 27th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of pre-cultures with B0034 (strong RBS), K577894 (Tet repressed YFP), KK577895 (Tet repressed RFP), F13+F14 === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Add 50 µL ampicillin(B0034)/chloramphenicol(K5777894, K577895, F13+F14) to 50 mL LB-medium | ||
| + | * Picking colonies from plate (XL1 blue; all rescue) | ||
| + | * Incubate over night at 37°C | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Production of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ampicillin (2 mL) was added to the Medium (1:1000) | ||
| + | * The pre-culture (50 mL) was poured into the Medium | ||
| + | * Culture was incubated at 37°C and 140 rpm until OD550 reached 0.5 | ||
| + | * To induce streptavidin expression anhydro-tetrazycline (200 µL) was added to the culture (1:10000) | ||
| + | * The culture was incubated at 37°C and 140 rpm for 4 hours | ||
| + | |||
| + | '''Result:''' | ||
| + | Streptavidin expression by BL21 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Re-sequencing of P30 (Tet repressed GFP in pSB1C3) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Purification and sequencing of P30 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * P30 was heated up to 100 °C and centrifugated afterwards to remove proteins from Promotor (if there are some), the supernatant is sequenced | ||
| + | * Sequencing batches were prepared after manufacturer's protocol | ||
| + | |||
| + | *(15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer) | ||
| + | *The different plasmids we prepared received the following barcodes: | ||
| + | **P30 (VF2) : FR11326638 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium === | ||
| + | |||
| + | '''Investigator: JH, NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of Streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4°C, 5000 rpm, 20 mins, F 4X1L rotor) | ||
| + | * The supernatant was cast away and the pellet was stored at –20 °C | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
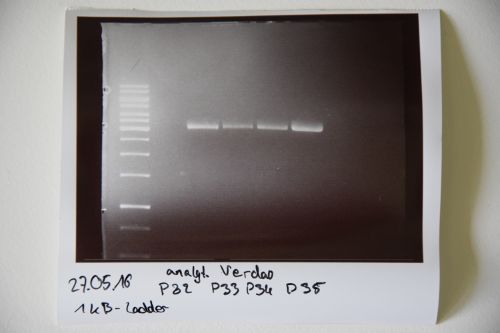
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' MiniPrep of cloned pSB1C3-A3C5 and –AviTag Plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MiniPrep of inoculated precultures was performed according to manufacturer’s protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * Concentrations were measured (in ng/µL): | ||
| + | |||
| + | Colony 1 of pSB1C3- AviTag P32: 120 | ||
| + | Colony 2 of pSB1C3- AviTag P33: 110 | ||
| + | Colony 1 of pSB1C3-A3C5 P34: 126 | ||
| + | Colony 2 of pSB1C3-A3C5 P35: 188 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: NA, JH, CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Initial vector digestion was performed with NgoMiV and AgeI, producing complementary overhangs. Religated pSB1C3 (due to incomplete vector dephosphorylation) loses the AgeI recognition sequence. Therefore plasmids without insert should run in the supercoiled position at about 1.5 kb, plasmids with inserts should run in the linearised position at 3 kb. Single enzyme digestion was chosen, because the insert is too small to be solved on a gel. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * analytic digestion: 3 µl of plasmid DNA P32 to P35 (about 300 ng) were digested with 0.3 µl AgeI-HF in 1 µl CutSmart Buffer and 5.7 µl H2O. The reaction was incubated for 2 h at 37°C. | ||
| + | * 5 µl digestion mix were loaded on a 1% agarose gel for electrophoresis, 1 kb ladder | ||
| + | |||
| + | '''Result:''' | ||
| + | *P32 to P35 might carry the insert because they all appear in the linearized position at 3 kb | ||
| + | *Therefore plasmid sequencing should confirm the result. | ||
| + | |||
| + | Lane 1: 1 kb ladder | ||
| + | Lane 3: P32 | ||
| + | Lane 4: P33 | ||
| + | Lane 5: P34 | ||
| + | Lane 6: P35 | ||
| + | |||
| + | [[File:Muc16_Cloning_of_A3C5_and_Avi-Tag_into_pSB1C3.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 3 (May 30th - June 5th)= | ||
| + | <div class="week" id="Week_3"> | ||
| + | |||
| + | =='''Monday, May 30th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Sequencing of P32 and P35 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *Sequencing batches were prepared according to manufacturers protocol (Mix2Seq Kit, Eurofins Genomics): 15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer (10 µM) | ||
| + | |||
| + | *The plasmids received the following barcodes: | ||
| + | **P32 (VF2) : FR11326637 | ||
| + | **P35 (VF2) : FR11326636 | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Religation of F13 (digest of pSB1C3 with AgeI and NgoMIV) and transformation in XL1 blue === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Re-ligate digested and dephosphorylated pSB1C3 and transformation in XL1blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Plasmid was ligated with the T4 DNA Ligase following the manufacturer’s protocol (NEB) | ||
| + | * 52 ng Vector DNA were used | ||
| + | * The Ligation mix was incubated for 30 min at RT | ||
| + | * 7 μL Ligation mixed were used for the transformation of 50 μL XL1 blue. | ||
| + | * Transformation was performed following the SOP | ||
| + | |||
| + | '''Result:''' | ||
| + | *5 clones on plate | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of pre-cultures with B0034 (strong RBS), K577894 (TET repressed YFP), KK577895 (Tet repressed RFP), F13+F14 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Add 50 µL ampicillin(B0034)/chloramphenicol(K5777894, K577895, F13+F14) to 50 mL LB-medium | ||
| + | * Picking colonies from plate (XL1 blue; all rescue) | ||
| + | * Incubate over night at 37°C | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, May 31th'''== | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Dialysis of eGFP === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Changing the buffer of eGFP for biotinylation (from TRIS to PBS) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * eGFP was poured in a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in ice cold PBS pH 7.4 | ||
| + | * The dialysis took place at 4°C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium === | ||
| + | |||
| + | '''Investigator: JB, NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of Streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The pellet was transferred into a beaker of sufficient size and resuspended in fridge-cooled Tris Buffer B (50 mM Tris/HCl (pH = 8.0), 1 mM EDTA) | ||
| + | * The solution was homogenized in the PANDA (see SOP) | ||
| + | * The resulting lysate was transferred into centrifuge tubes and spun down (4°C, 18,000 rpm, 30 mins, SS34-rotor). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Transformation of P30 (Tet repressed GFP)=== | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' expression of tet-repressed GFP in E. Coli BL21 | ||
| + | |||
| + | '''Procedure:''' transformation in competent E. Coli BL21 according to protocol | ||
| + | |||
| + | '''Result:''' plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
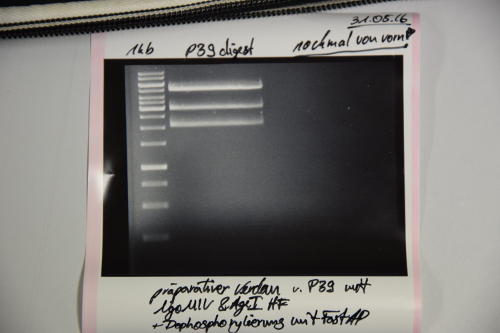
| + | <div class="receptor"> | ||
| + | === Cloning of A3C5 and Avi-Tag into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Preculture of pSB1C3 (F13+F14) was red | ||
| + | See if RFP-generator is still inside the pSB1C3 | ||
| + | See if ligation of EspP was successful | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MiniPrep of P39 was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * pSB1C3 was digested with AgeI and NgOMIV (1.5 µL P39 + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 22.5 µL ddH2O; incubation for 2h, 37°C) | ||
| + | * FastAP was added and the reaction was incubated for another 10min at 37°C | ||
| + | * FastAP was inactivated by heating the mix to 75°C for 10min. | ||
| + | * The digestion was purified by gelelctrophoresis (1%, 75V, 1h) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_P39_digest.png|500px]] | ||
| + | |||
| + | Unknown signals. | ||
| + | Repeat transformation, digestion, dephosphorylation and ligation of pSB1C3 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Re-transformation of P4 (pSB1C3 (RFP-generator)) in XL1 blue === | ||
| + | |||
| + | '''Investigator: JH, CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2 μL P4 were used for transformation of XL1 blue | ||
| + | * Transformation was performed according to SOP (incl. rescue plate) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Competent E. coli XL1 cells === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Generation of competent E.coli XL1 cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 ml LB w.o. antibiotics (sterile) | ||
| + | * Inoculation of one colony of E.coli XL1 cells | ||
| + | * Incubate overnight at 37°C | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 1th'''== | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === eGFP Biotinylation === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Biotinylation of dialysed eGFP past IEC | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The average concentration of the pooled factions 12 and 13 should be about 0.5 mg/ml (=19 µM) | ||
| + | * 10 ml of a 100 mM Biotin stock-solution was prepared (340 mg) | ||
| + | * 3.5 ml of eGFP were biotinylated with 12 µl 100 µM Biotin-solution (20x molar excess) | ||
| + | * Incubation at 4 °C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === pSB1C3 vector backbone preperation === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Inoculation of two colonies pSB1C3-RFP Generator | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2x 5 ml LB+Cam medium (1:1000) | ||
| + | * Picking colonies from plate (XL1 blue, rescue) | ||
| + | * Incubate overnight at 37°C | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Generation of GFP expressing E. coli cells === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Inoculation of BL21 transformed cell with P30 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 50 ml LB+Cam medium (1:1000) | ||
| + | * Picking colonies from plate (BL21, rescue) | ||
| + | * Incubate over night at 37°C | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Competent E. coli XL1 cells === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Generation of competent E. coli XL1 cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculated 500 µl preculture in 50 ml LB media w.o. antibiotics | ||
| + | * Incubated and shaken at 37°C until the OD550 reached 0.44 | ||
| + | * Transferred into a falcon tube and incubated on ice for 10 mins | ||
| + | * Spun down at 3000 g for 10 mins at 4°C, supernatant cast away | ||
| + | * Pellet resuspended in 40 ml fridge-cooled MgCl2 (100 mM), once again spun down at 4°C for 10 mins at 3000 g | ||
| + | * Supernatant cast away, pellet resuspended in 20 ml cold CaCl2 (50 mM) | ||
| + | * Incubation on ice for 30 mins | ||
| + | * Centrifugation repeated, supernatant cast away | ||
| + | * Pellet resuspended in 2 ml of CaCl2 (50 mM) and glycerol (15%)-solution, aliquoted (50 µl each), frozen in liquid nitrogen and stored at -80°C for further use | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of Pcat-RBS-construct in pSB1A2 === | ||
| + | |||
| + | '''Investigator: CG, JH, CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Pcat (I14033) was cut out of P27 and ligated into P38 for generating the Pcat-RBS (B0034) construct. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * P27 was digested with EcoRI and SpeI (4 µL P27 + 1.5 µL EcoRI + 1.5 µL SpeI + 3 µL CutSmart + 20 µL ddH2O; incubation for 2h, 37°C) | ||
| + | * P38 was digested with EcoRI and XbaI (10 µL P27 + 1.5 µL EcoRI + 1.5 µL XbaI + 3 µL CutSmart + 14 µL ddH2O; incubation for 2h, 37°C) | ||
| + | * Preperative gel electrophoresis was performed | ||
| + | * For P27 the signal at about 50 bp was cut out and gel extracted according to manufacturer's protocol (Gelextractionkit, Qiagen): 5.9 ng/µl | ||
| + | * For P30 the signal at about 2.1 kb was cut out and gel extracted according to manufacturer's protocol (Gelextractionkit, Qiagen): 8.9 ng/µl | ||
| + | * Ligation according to SOP | ||
| + | </div> | ||
| + | <div class="streptavidin"> | ||
| + | === Purification of streptavidin === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Refolding denaturized streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * the GdmCl-solved streptavidin was spun down using an SS-34 rotor (~60 min, 18,000 rpm, 4°C) | ||
| + | * The supernatant containing the solved and denaturized protein was transferred into a falcon tube, the pellet cast away | ||
| + | * The protein solution was slowly dripped into 4 l of cooled 1x PBS (in the 4°C room) using a hydraulic pump and stirred overnight | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 2th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Preparation of pSB1C3 for further use === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of pSB1C3 to remove RFP-generator | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * P40 was digested with AgeI and NgOMIV (2.5 µL P40 + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 21.5 µL ddH2O) | ||
| + | * Incubation over night at 37°C | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Generation of GFP expressing E. coli cells === | ||
| + | |||
| + | '''Investigator: CG, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Various inductor concentrations | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 12x 5 ml LB+Cam media were inoculated with 50 µl preculture | ||
| + | * when cultures reached OD550 of 0.5, they were induced with tetrazycline-anhydride of various concentrations (0 ng/µl, 20 ng/µl, 40 ng/µl etc., 200 ng/µl) | ||
| + | * t=0, =30, =90, =150, =210, =270 a 100 µl aliquot of each culture was stored at 4 °C in the refrigerator | ||
| + | </div> | ||
| + | <div class="polymerization"> | ||
| + | === Dialysis of biotinylated eGFP === | ||
| + | |||
| + | '''Investigator: JH, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of biotinylated eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * eGFP was poured in a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in 2 L ice cold Tris/HCl 20 mM pH 8.0 | ||
| + | * The dialysis took place at 4°C over night | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Ligation of F17.1 (p(cat)) and F18 (stron RBS) and transformation in XL1 blue === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Ligation and transformation in XL1blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Plasmid was ligated with the T4 DNA Ligase following the manufacturer’s protocol (NEB) | ||
| + | * 9 µL F18 (B0034 in pSB1A2; RBS) and 1 µL of F17.1 (I14033; P(cat)) were used [AmpR] | ||
| + | * The Ligation mix was incubated for at least 2h at RT | ||
| + | * 7 μL Ligation mixed were used for the transformation of 50 μL XL1 blue | ||
| + | * Transformation was performed following the SOP [AmpR], incl. rescue plate | ||
| + | </div> | ||
| + | |||
| + | =='''Friday, June 3th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of F8 (A3C5), F9 (Avi-Tag) and F14 (EspP) into F20 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Cloning of A3C5, Avi-Tag and into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * FastAP was added and the digestion of F20 and was incubated for another 15 min at 37°C | ||
| + | * The digestion was purified by gelelctrophoresis (1%, 75V, 1h) | ||
| + | * Plasmid backbone (signal 1) was cut from the gel | ||
| + | * Gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_digestion_F20.png|500px]] | ||
| + | |||
| + | Signal 1: F20 | ||
| + | |||
| + | [F20]= 13,8 ng/μL | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Generation of GFP expressing E. coli cells === | ||
| + | |||
| + | '''Investigator: CG, NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' SDS PAGE for verification of successful GFP expression | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * SDS electophoresis according to SOP | ||
| + | * Lane 1: #12, 90 min | ||
| + | * Lane 2: #12, 150 min | ||
| + | * Lane 3: #12, 210 min | ||
| + | * Lane 4: #12, 270 min | ||
| + | * Lane 5: 8 µl | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Generation_of_GFP_expressing_E_coli_cells.png |500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of colonies from F19 (pcat-strong RBS)=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' three colonies were picked from yesterdays transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3x 5 ml LB+Amp media | ||
| + | * Each culture was inoculated with one colony | ||
| + | * Incubation at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of F8 (A3C5), F9 (Avi-Tag) and F14 (EspP) into F20 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F21, F22 and F23 into F20 and transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ligation was performed according to the SOP with: 0.7 µl F8 (1:10) / 9.3 µl F20, 0.7 µl F9 (1:10) / 9.3 µl F20, 9.1 µl F14 / 0.9 µl F20 | ||
| + | * Transformation was performed according to the SOP on LB Cam-Agar | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Precipitation of Streptavidin === | ||
| + | |||
| + | '''Investigator: NA, JB, LK''' | ||
| + | |||
| + | '''Aim of the Experiment:''' Purification of Streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Slowly add (NH4+)2SO42- until the concentration reaches 40% (23,1 g/100mL) while stirring | ||
| + | * Let it stir over night to ensure complete precipitation | ||
| + | * Centrifuge at 8500 rpm at 4°C for 15min (rotor: SLA1500), dismiss the pellet and use the supernatant for further processing | ||
| + | * Slowly add (NH4+)2SO42- until the concentration reaches 70% (19,1 g/100mL) while stirring | ||
| + | * Let it stir over night (slowly to avoid foaming) | ||
| + | * Centrifuge at 8500 rpm at 4°C for 15min (rotor: SLA1500), dismiss the supernatant and use the pellet for further processing | ||
| + | * Resuspend the protein pellet in a minimum volume of TRIS/HCl; pH 8; 20mM (NO SALT) (6 mL per pellet), the pellets where then united and titrated with Buffer until the solution became clear. Total amount of buffer: 40mL | ||
| + | * Transfer to 50 mL Falcon Tubes and centrifuge at full speed for 15 min. Filter the supernatant through 45nm sterile filter and keep the pellet (rescue). | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, June 4th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | ===Inoculation of colonies from F21 (pSB1C3 (A3C5)), F22 (pSB1C3 (Avi-Tag)), F23 (pSB1C3 (EspP)) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' three colonies were picked from each transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3x 5 ml LB+Cam media for each fragment | ||
| + | * Each culture was inoculated with one colony | ||
| + | * Incubation at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === MiniPrep of F19 (pSB1A2 (pcat-strong RBS))=== | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extraction of Ligation: F17+F18 (starke RBS+P(cat)) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | Concentrations: K1= 66,2 ng/µl; K2= 60,2 ng/µl; K3= 104,4 ng/µl | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 4 (June 6th - June 12th)= | ||
| + | <div class="week" id="Week_4"> | ||
| + | |||
| + | =='''Monday, June 6th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of TetR from p28 into p31 (pSB1C3(weak RBS)) === | ||
| + | |||
| + | '''Investigator: LK, JH, NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Cloning TetR in front of weak RBS in pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestion of p28 with EcoR1 and Spe1 (20µl DNA, 2,5 µl each enzyme, 5µl CutSmart buffer, 20µl ddH2O) and p31 (4µl DNA, 1µl EcoRI / XbaI, 2µl, 12µl DNA) -> Incubation at 37°C for 2h. | ||
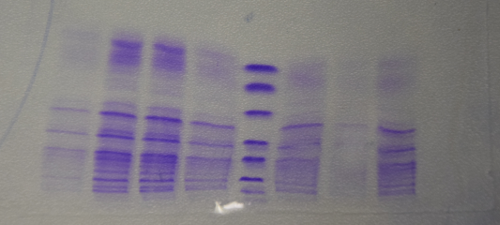
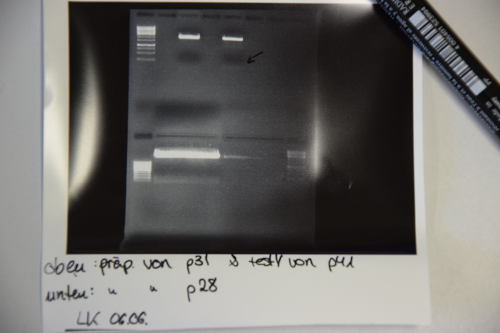
| + | * Electrophoresis: 1% Agar, 15 min , 70 V (oben p31 und p41, unten p28) | ||
| + | |||
| + | [[File:Muc16_praep_P31_p28.png|500px]] | ||
| + | |||
| + | * Gelextraction concentrations: p28 (F25) = 2,3 ng/µl | ||
| + | p31 (F24) = 7,8 ng/µl | ||
| + | * Ligation with 6,3 µl vector (F24) and 1,7 µl Insert (F25) + 1µl Buffer + 1µl Ligase over night, 16°C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
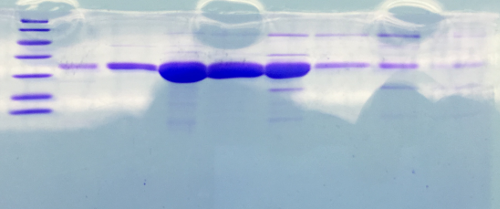
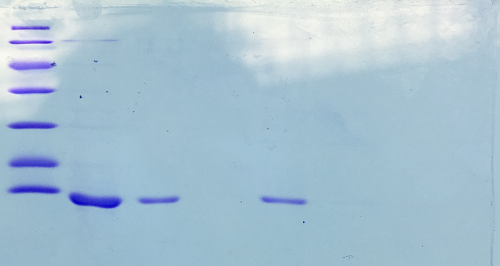
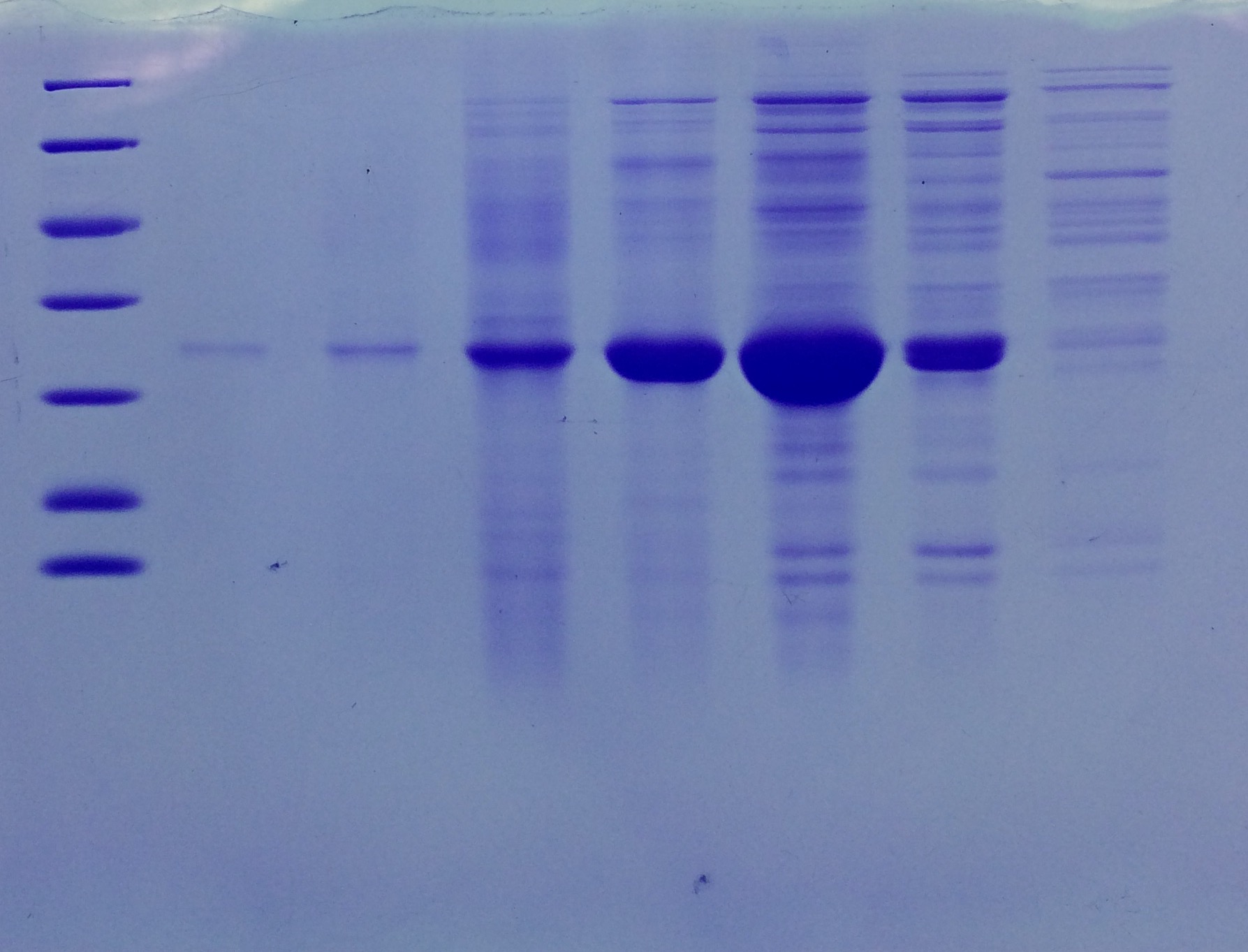
| + | === SDS-PAGE of Streptavidin === | ||
| + | |||
| + | '''Investigator: NA, JH, LK''' | ||
| + | |||
| + | '''Procedure:''' mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining. Concentration of streptavidin was measured via UV/VIS spectroscopy. | ||
| + | |||
| + | c = 23,5 mg/ml | ||
| + | |||
| + | '''Result:''' | ||
| + | |||
| + | * 1. Lane: 8 µl Marker | ||
| + | * 2. Lane: 5 µl of Streptavidin_1:10 | ||
| + | * 3. Lane: 5 µl of Streptavidin_1:50 | ||
| + | * 4. Lane: 5 µl of Pellet_1:500 | ||
| + | * 5. Lane: 5 µl of after 40% precipitation_1:10 | ||
| + | * 6. Lane: 5 µl of supernatant after 70% precipitation and centrifugation | ||
| + | |||
| + | Successful Streptavidin production verified. | ||
| + | |||
| + | [[File:SDS-PAGE_of_Streptavidin.png |500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P41 (pSB1C3 (strongRBS-p(cat))) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P41 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (100 ng/ µl) and 2 µL sequencing primer (VF2)) | ||
| + | |||
| + | The plasmids prepared received the following barcode: | ||
| + | * P41 : FR11326633 | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 7th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of P30 (Tet repressed GFP), P36 (Tet repressed RFP), P37 (Tet repressed YFP) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of RFP, GFP and YFP in BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. Coli BL21 according to protocol + rescue | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
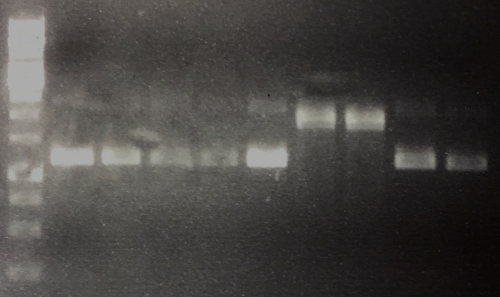
| + | === MiniPrep of P42 to P50, analytic digestion, sequencing === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | |||
| + | * Concentrations (ng/µl each) for P42: 260, P43: 247, P44: 181, P45: 118, P46: 261, P47: 135, P48: 148, P49: 240, P50: 299 | ||
| + | * Analytic digestion: 0.3 µl AgeI, 2 µl CutSmart, 6.7 µl H2O for P42, P43, P44, P45, P46, P49, P50 and 0.3 µl EcoRI, 0.3 µl PstI, 2 µl CutSmart, 6.4 µl H2O for 2 h at 37°C | ||
| + | * 1% agarose gel electrophoresis | ||
| + | * Sequencing: P42= FR11326632 (VF2), P45 =FR11326631 (VF2) | ||
| + | |||
| + | '''Results:''' | ||
| + | * Lane 1: 5 µl 1 kb Ladder | ||
| + | * Lane 2-10: Digestion of P42 to P50. P47 and P48 are carrying the insert. All other plasmids are religated and lost their AgeI site, hence they run in the ccc position. | ||
| + | |||
| + | [[File:Muc16_miniPrep_of_P42_to_P50.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Dialysis of not biotinylated eGFP === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of eGFP for mass determination | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * eGFP was poured in a dialysis tubes | ||
| + | * The tubes were then placed in ice cold ammonium acetate | ||
| + | * The dialysis took place at 4°C over night | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Polymerisation attempts === | ||
| + | |||
| + | '''Investigator: JH, CR, CG, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Identification of optimal polymerisation conditions for biotinylated eGFP and BSA in a Streptavidin reservoir | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * variate of compositions for both solutions was examined to modify density properties during “printing” | ||
| + | * all compositions can be found in excel sheet | ||
| + | |||
| + | '''Results:''' | ||
| + | * no or little successful polymerization at Streptavidin concentrations of 4 mg/mL and below; immense density incompatibilities | ||
| + | * sucessfull polymerization at Streptavidin concentrations of 10 mg/mL and BSA solution with approx. 15% glycerol | ||
| + | * no success with biotinylated eGFP (too few Biotin molecules?!) | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 8th'''== | ||
| + | |||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P36 (Tet repressed RFP) and P37 (Tet repressed YFP) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P36 and P37 in both directions | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | |||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | * P36 (VF2): FR11326630 | ||
| + | * P36 (VR): FR11326629 | ||
| + | * P37 (VF2): FR11326628 | ||
| + | * P37 (VR): FR11326627 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Dialysis of Streptavidin === | ||
| + | |||
| + | '''Investigator: NA, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of Streptavidin (possible ammonium sulfate contamination after precipitation) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Streptavidin was poured in a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in 3 L ice cold Tris/HCl 20 mM pH 8.0 (no salt) | ||
| + | * The dialysis took place at 4°C over night | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Polymerisation attempts === | ||
| + | |||
| + | '''Investigator: JH, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Identification of optimal polymerisation conditions for biotinylated BSA in a Streptavidin reservoir | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * variate of compositions for both solutions was examined to modify density properties during “printing” (Streptavidin at 10 mg/mL or higher) | ||
| + | * all compositions can be found in excel sheet | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, June 9th'''== | ||
| + | <div class="general"> | ||
| + | === Generation of GFP, YFP, RFP expressing E. coli cells === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Various inductor concentrations, various fluorescent proteins were expressed in E coli cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3x 5 ml LB+Cam media were inoculated with 50 µl preculture of GFP (P30), RFP (P36), YFP (P37) | ||
| + | * when cultures reached OD550 of 0.5, they were induced with anhydro-tetrazycline of various concentrations (0 ng/µl, 100 ng/ml, 200 ng/ml) | ||
| + | * 50 ng/ml = 12,5 µl of 1:100 2 mg/ml stock solution, 200 ng/ml = 50 µl of 1:100 2 mg/ml stock solution | ||
| + | * t=0, =4 h and stored on ice | ||
| + | |||
| + | '''Resuts:''' no fluorescent could be detected. Therefore an overnight induction at 37°C with 1 µM/ml inductor concentration was performed with the RFP plasmid. | ||
| + | </div> | ||
| + | <div class="polymerization"> | ||
| + | === Biotinylation of GFP expressing E. coli cells === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Biotinylation of P30 carrying E. coli cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 850 µl P30 preculture from Thursday 2nd were centrifuged and the supernatend was discarded | ||
| + | * Pellet was resuspended in 800 µl PBS-buffer | ||
| + | * 100 µl 100 mM Biotin-NHS was added | ||
| + | * incubation for 4 h at room temperature | ||
| + | * Experimenting with some glycerol and streptavidin concentrations. A more structured approach will be done the next day. | ||
| + | |||
| + | '''Results:''' A shape thin “sausage” could be detected. Therefore a bigger E. coli culture was biotinylated at 4°C overnight for experimenting with the next day. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of TetR promotor and weak RBS in pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' MiniPrep and NanoDrop of F26 K1, K2 and K3 | ||
| + | Sequencing of P51 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | Concentrations were as following: | ||
| + | * P51: 163,3 | ||
| + | * P52: 151,7 | ||
| + | * P53: 140,5 | ||
| + | |||
| + | P51 was sequenced: | ||
| + | * 1. with VF2: FR11326626 | ||
| + | * 2. with VR: FR11326625 | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Biotinylation of eGFP === | ||
| + | |||
| + | '''Investigator: VL, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Biotinylation of eGFP with 200x biotin-NHS molar excess over eGFP (15x excess over surficial K residues). NB: previous trial unsuccessful. Excess of biotin was likely insufficient to ensure efficient biotinylation, esp. since biotin-NHS slowly hydrolyzes in the aqueous environment. | ||
| + | |||
| + | '''Procedure:''' 1 ml of eGFP was biotinylated with an 200x excess of biotin in DMF (100 uM; 38 uL), 5.5 h at rt. The solution was dialysed versus 20 mM Tris/HCl pH 8 overnight at 4 °C. | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === PCR of gene syntheses === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification od iGEM_1_SA-m1 (F27), iGEM_1_mRuby_EGFR (F28) and iGEM_2_IRES-BirA (F29) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Add 100µl of ddH2O to the gene sythesis samples and incubate for 20 min at 50°C | ||
| + | * Dilute samples to a final concentration of max. 1ng/µl (1:10) with water. | ||
| + | * Approach: | ||
| + | |||
| + | 1µl DNA (diluted) | ||
| + | |||
| + | 5µl buffer | ||
| + | |||
| + | 0,5µl dNTPs | ||
| + | |||
| + | 1,25µl forward Primer VF2 | ||
| + | |||
| + | 1,25µl reverse Primer VR | ||
| + | |||
| + | 0,25µl Q5 Polymerase | ||
| + | |||
| + | Fill up with water up to 25µl | ||
| + | |||
| + | * Make sure to pipette everything on ice and add the polymerase at the end | ||
| + | * Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/) | ||
| + | |||
| + | 98°C_1min initiation step | ||
| + | |||
| + | 98°C_10sec | ||
| + | |||
| + | 66°C_30sec > step 2-4: 35 repeats | ||
| + | |||
| + | 72°C_30sec | ||
| + | |||
| + | 72°C_2min final step | ||
| + | |||
| + | 4°_hold | ||
| + | |||
| + | * The samples were then prepared with the PCR Product Purification Kit | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Polymerisation experiments for biotinylated eGFP and BSA with Streptavidin === | ||
| + | |||
| + | '''Investigator: JH, CR, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Determination of ideal concentrations for both protein interaction partners as well as buffer viscosity (glycerol content etc.) for polymerisation in a test tube | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Different concentrations of proteins and glycerol were tested to support stable polymerisation without protein aggregates rising to the surface or sinking to quickly | ||
| + | * The experiments done are not going to be further documented as the streptavidin used for the experiments was previously determined in a wrong fashion and was actually considerably lower than expected (4x lower). Streptavidin was, due to that circumstance, concentrated to be able to support experiments with at least 5 mg/ml concentrations | ||
| + | * First experiments with concentrated streptavidin seemed promising, though a complete series of experiments with varying parameters should be conducted in the future | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Sequencing of P29 (pSB1C3 (double terminator), P36 (pSB1C3 (Tet repressed RFP)) and P37 (pSB1C3 (Tet repressed RFP)) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P29, P36 and P37 | ||
| + | |||
| + | '''Procedure:''' According to the manufacturer's protocol. | ||
| + | * P29: FR11326624 (VF2) | ||
| + | * P36: FR11326623 (VF2) | ||
| + | * P37: FR11326621 (VR), FR11326622 (VF2) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Inoculation of (pre)cultures for a pSB1C3 midiprep === | ||
| + | |||
| + | '''Investigator: JB, VM''' | ||
| + | |||
| + | '''Aim of the experiment:''' Massively amplifying the pSB1C3 vector (which will be needed for building up new biobricks) by midiprepping the P40 plasmid (RFP-generator in pSB1C3). Similar to minipreps, but on a greater scale. | ||
| + | '''Procedure:''' 3 ml precultures were inoculated with a clone from the plate previously used for minipreps. After the precultures had grown for 4 hours (due to shortage of time), 2 x 50 ml cultures were inoculated and shaken overnight at 37°C. The next day, cultures were put into the fridge for storage until Monday. | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 5 (June 13th - June 19th)= | ||
| + | |||
| + | <div class="week" id="Week_5"> | ||
| + | |||
| + | =='''Monday, June 13th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Digestion of gene synthesis iGEM1 and iGEM2 === | ||
| + | |||
| + | '''Investigator: LK, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digest F28 (iGEM1) and F29 (iGEM2) in order to ligate them into P40, which was equally digested (to get rid of the rfp-generator and have pSB1C3 as backbone) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 30µl of F28 were digested for 3h with SpeI and NgoMIV, and 30 µl of F29 were digested with XbaI and PstI | ||
| + | * 2 x 20 µl where digested with the equivalent enzymes for 3h | ||
| + | * After electrophoresis the bands were cut out and purified | ||
| + | |||
| + | '''Results:''' Results show that PCR of F28, F29 might not have worked. P40 was extracted from gel (F30 and F31). PCR of gene synthesis was repeated (50µl!) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Midiprep of P40 === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of P40 from | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * See protocol of Midiprep kit | ||
| + | * 1xuse-centrifugation-columns broke in centrifuge during last step, DNA rescue was tried by resuspending rest of the solution in the tube in EB buffer and redo the last steps (isoprop and ethanol precipitation) | ||
| + | * Airdry "pellet"(no pellet visible) for 15 min under fume hood and resuspend in 100µl EB Buffer | ||
| + | * Measure concentration with nanodrop | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Preparation of Collagen + Biotinylation === | ||
| + | |||
| + | '''Investigator: VL, JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' prepare collagen solution + biotinylate small sample | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Dissolve 5 g collagen in 50 mL Tris/HCl pH 8 20 mM w/o salt --> 770 uM solution (100 mg/mL) | ||
| + | * Sterile filter | ||
| + | * Biotinylation: 980 uL Tris buffer + 20 uL collagen solution + 30 uL biotin-NHS (100 mM in DMF, 200x molar excess over collagen monomers) | ||
| + | * Reaction time: | ||
| + | * Dialyze 4 °C Tris/HCl pH 8 20 mM w/o salt o/n | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Transformation of P9 (pASK75) === | ||
| + | |||
| + | '''Investigator: JL, VL''' | ||
| + | |||
| + | '''Aim of the experiment:''' transform streptavidin expression vector P9 (clone 4) into E. coli BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Standard (see booklet): 45 s at 42 °C; incubate 1h, 37°C; plate: medium w. chloramphenicol (+ rescue plate) Incubate o/n 37°C. | ||
| + | * Wrong antibiotic plate, repeated the next day of the experiment:''' Massively amplifying the pSB1C3 vector (which will be needed for building up new biobricks) by midiprepping the P40 plasmid (RFP-generator in pSB1C3). Similar to minipreps, but on a greater scale. | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 14th'''== | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Mass spectrometry of biotinylated eGFP === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' identify biotinylation degree of eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | [generic_display_report] (i could not upload this file, sorry.) | ||
| + | |||
| + | '''Results:''' about 4-5 biotins per protein. To verify the biotinylation degree the unbiotinylated eGFP will be measured. | ||
| + | </div> | ||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of PCR products | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Volume of PCR reaction: 50 µL | ||
| + | * PCR products were purified by manufacturers protocol (QIAquick PCR purification kit, Qiagen) | ||
| + | * Concentration measured by Nanodrop: | ||
| + | * F32: 87,5 ng/µL | ||
| + | * F33: 68,3 ng/µL | ||
| + | * Test if PCR worked: | ||
| + | * Samples were applied on a 2% agarose gel and was run at 80 V for 40 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_PCR_F32_F33.png|500px]] | ||
| + | |||
| + | * PCR of F28 worked --> F32 | ||
| + | * PCR of F29 did not work --> wrong primers were used for amplification --> F33 not right (Re-Do PCR with appropriate primers | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of F32 (SpeI, NgoMIV) and F33 (XbaI, PstI) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestion F32: 27 µl of F32 (2.4 µg), 5 µl CutSmart Buffer, 1.5 µl SpeI, 2 µl NgoMIV, 14.5 µl H2O | ||
| + | * Digestion F33: 27 µl of F32 (1.8 µg), 5 µl CutSmart Buffer, 1.5 µl XbaI, 1.5 µl PstI, 15 µl H2O | ||
| + | * Incubation for 3 h at 37°C | ||
| + | * Preperative gel: | ||
| + | |||
| + | [[File:Muc16_PCR_F32_F33_praep-gel.png|500px]] | ||
| + | |||
| + | * Gelextraction of F32 upper band and F32 lower band was executed by manufacturers protocol (Qiagen) | ||
| + | * F32 upper band (mRuby) was named F34 (=10 g/yl) | ||
| + | * F32 lower band (eGFR) was named F35 (=6 ng/yl) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F34 (mRuby) and F35 (eGFR) in F30 (pSB1C3); Tranformation in XL1blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ligation mixtures were prepared as following: | ||
| + | * F34: 8.0 μL F34, 2 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase) | ||
| + | * F35: 7.6 μL F35, 2.4 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase) | ||
| + | * Ligation was incubated for 30 min at RT | ||
| + | |||
| + | [something is missing here] | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Re-Sequencing of P37 (Tet repressed YFP) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification and sequencing of P30 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * P30 was heated up to 100 °C and centrifugated afterwards to remove proteins from Promotor (if there are some), the supernatant is sequenced | ||
| + | * Sequencing batches were prepared after manufacturer's protocol | ||
| + | * (15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer) | ||
| + | * The plasmid we prepared received the following barcode: P37 (VF2) : FR11326620 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Streptavidin expression === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Transformation of P9 (pASK74 (SA1)) in BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation was executed following SOP | ||
| + | * Transformed cells were plated on LB medium (Amp) (50 µL and rescue (50 µL) | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Dialysis of Streptavidin and biotinylated BSA === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Streptavidin and bitinylated BSA are both in Tris/HCl without salt. SInce we will perform later experiments in salty solutions the dialysis was performed against 137 mM NaCl (Saltconcetration of PBS) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 L Tris/HCl NaCl solution was prepared | ||
| + | * Dialysis was prepared according to SOP | ||
| + | * Cut off Dialysis hose: 14 kDa | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Inoculation of (pre)cultures of P40 for a pSB1C3 midiprep [repeat] === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Massively amplifying the pSB1C3 vector by midiprepping the P40 plasmid (RFP-generator in pSB1C3) | ||
| + | |||
| + | '''Procedure:''' four 4 ml precultures were inoculated with a clone from the plate previously used for minipreps. After the precultures had grown for 6 hours, 50 ml cultures were inoculated and shaken overnight at 37°C. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Subcloning of P41 (strong RBS-p(cat)) Insert into pSB1C3 === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Moving P41 (pSB1A2; Pcat+weakRBS) to pSB1C3 for biobrick submission | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestion of P41 with XbaI and PstI (10 µl DNA, 1,5 µl each enzyme, 3 µl CutSmart buffer x10, 14 µl ddH2O) -> Incubation at 37°C for 2h. | ||
| + | * Electrophoresis: 2% Agar, 40 min , 90 V (1kb ladder, empty, P41, 100b ladder) | ||
| + | * GelExtraction | ||
| + | |||
| + | '''Resuts:''' no visible signal of insert at approx. 100b. Area was cut blind for GelEx. No DNA in the sample according to NanoDrop measurement. Repeated the next day. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of colonies from NanoLuc === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2x 4 ml LB+Cam media | ||
| + | * Each culture was inoculated with one colony | ||
| + | *Incubation at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 15th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Subcloning of P41 (strong RBS-p(cat)) Insert into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Moving P41 (pSB1A2; Pcat+weakRBS) to pSB1C3 for biobrick submission. No gel extraction was performed because the fragement is too small to visualize on the gel. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestion of P41 with XbaI and PstI (10 µl DNA, 1,5 µl each enzyme, 3 µl CutSmart buffer x10, 1 µl FastAP, 13 µl ddH2O) -> Incubation at 37°C for 2 h, heat inactivation at 80°C for 20 min. | ||
| + | fragement is to small to visualize on agel. | ||
| + | * Subcloning into F31: 3.8 μL F38, 6.2 μL F31, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase according to the SOP | ||
| + | * Transformation into E. coli XL1 according to the SOP | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Miniprep of pSB1C3(Nanoluc) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Preperation of a BioBrick with NanoLuciferase in pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *The Miniprep was done according to the protocol. | ||
| + | *The two tubes where pooled. | ||
| + | *The new Plasmid is found under P54 and was sent to sequencing. | ||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | === Polymerisation === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Determine optimal polymerisation conditions for BSA and Collagen | ||
| + | |||
| + | '''Procedure:''' The experiment was performed in a 96 Well plate: | ||
| + | |||
| + | [[File:Muc16_excel_table.png|500px]] | ||
| + | |||
| + | [[File:Muc16_well_plate.png|500px]] | ||
| + | |||
| + | '''Results:''' maximum concentration of Streptavidin and biotinylated Protein works best. Collagen does not work. Glycerolconcentrations do not inhibit interaction. Reaction might be to slow at given concentrations to be effective in printing process. | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MidiPrep of pSB1C3 with RFP generator (P4) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of pSB1C3 with RFP generator for further processing (P56) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | Concentration very low (12ng/μL), but high volume (500 μL) | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, June 16th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === MiniPrep of F36 (Ligation mRuby in pSB1C3) K1 & K2, F37 (Ligation EGFR in pSB1C3), I13453 (pBad promotor) K1 & K2, K577881 (pC+AraC+pBad+GFPc) K1 & K2 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' isolate Plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * Plasmids were named as following: [concentration] | ||
| + | |||
| + | F36 K1: P57 [212,9 ng/μL] | ||
| + | |||
| + | F36 K2: P58 [181,3 ng/μL] | ||
| + | |||
| + | F37: P59 [195,5 ng/μL] | ||
| + | |||
| + | K577881 K1: P60 [557,2 ng/μL] | ||
| + | |||
| + | K577881 K2: P61 [791,9 ng/μL] | ||
| + | |||
| + | I13453 K1: P62 [130 ng/μL] | ||
| + | |||
| + | I13453 K2: P63 [460 ng/μL] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Sequencing of P29 (double terminator), P36 (Tet repressed RFP) and P37 (Tet repressed YFP)=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P56, P57, P59, P61 and P62 | ||
| + | |||
| + | '''Procedure:''' According to the manufacturer's protocol. | ||
| + | |||
| + | * P56: FR11326618 (VF2) | ||
| + | * P57: FR11326617 (VF2) | ||
| + | * P59: FR11326616 (VF2) | ||
| + | * P61: FR11326613 (VR), FR11326615 (VF2) | ||
| + | * P63: FR11326614 (VF2) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MidiPrep P4 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: VL''' | ||
| + | |||
| + | '''Aim of the experiment:''' isolate Plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen). Centrifugation steps at 20,000 g performed at 12,500 rpm in special centrifugation tubes. Since only one column was left, both prepped cell pellets were applied to one column. Flow-through and wash was collected. | ||
| + | * Conc: | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Amplification of BioBricks (overnight cultures for minipreps) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Inoculation of 5 ml overnight cultures for minipreps. The following biobricks were used: | ||
| + | |||
| + | * BBa_K157012 | ||
| + | * BBa_K243004 | ||
| + | * BBa_K243006 | ||
| + | * BBa_K157001 | ||
| + | * BBa_K243005 | ||
| + | * BBa_I712009 | ||
| + | * BBa_K404108 | ||
| + | * BBa_K782061 | ||
| + | * BBa_K1371012 | ||
| + | * BBa_K782060 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Three hours of incubation of the tubes containing the bacterial agar at 37°C (Note: 12 hours are actually recommende) | ||
| + | * Inoculation of two times 5 ml preculture containing the appropriate antibiotic per clone. For KanRAmpR (reistance against both ampicillin and kanamycin) it was only selected for ampicillin. | ||
| + | * Incubation shaking overnight at 37°C | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR Amplification of genesynthesis 2 (F29; IRES-BirA) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of Genesynthesis_2 (IHRES_BirA_StrepTag) | ||
| + | |||
| + | '''Procedure:''' A 50µl approach was made: | ||
| + | * 33,5 µl Water | ||
| + | * 2µl DNA (F29, diluted 1:10) | ||
| + | * 2,5µl of each Primer (O11 and O12 c:10µM) | ||
| + | * 0,5µl dNTPmix | ||
| + | * 5µl Reaction buffer | ||
| + | * 0,5µl Q5-Polymerase | ||
| + | * The Annealing Temp. was set an 58°C | ||
| + | * Next step is to run an analytic gel (band should be visible at 1,5 kB) | ||
| + | </div> | ||
| + | |||
| + | =='''Friday, June 17th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of genesynthesis 2 (IRES-BirA) into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CG, NA, JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification oft he PCR reaction, gel verification, restriction digestion (F29), ligation with F31 and transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Yesterdays PCR was purified according to Qiagen PCR purification kit | ||
| + | * An anlaytic gel electrophoresis was performed on a 1% gel with 4 µl purified PCR product | ||
| + | * Restriction digest: 26 µl purified PCR product, 1.5 µl XbaI, 1.5 µl PstI, 5 µl NEB SmartCut, 16 µl H2O. Incubation for 2 h at 37°C. | ||
| + | * Gel extraction | ||
| + | * Ligation: F31 | ||
| + | * Transformation | ||
| + | |||
| + | '''Results:''' Gel electrophoresis after PCR purification exhibit a thin signal at 1.5 kb. Next time 1:30 min for elongation time might increase the signal. | ||
| + | |||
| + | [[File:Cloning_of_gene_synthesis_2_into_pSB1C3.png|200px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Miniprep new iGEM BioBricks === | ||
| + | |||
| + | '''Investigator: VL''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purify plasmid DNA from transformed cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Miniprep according to manual (pellet centrifuged 5000 rpm, 10 min, 4 °C) | ||
| + | * Elution with 50 uL EB Buffer | ||
| + | * Concentrations (all ng/uL): | ||
| + | |||
| + | K782061 (a) 172.9, (b) 53.0 | ||
| + | |||
| + | K243006 (a) 241.1, (b) 58.6 | ||
| + | |||
| + | K1371012 (a) 138.0, (b) 43.0 | ||
| + | |||
| + | K157001 (a) 184.3, (b) 186.4 | ||
| + | |||
| + | K782060 (a) 339.0, (b) 254.0 | ||
| + | |||
| + | K243005 (a) 62.3, (b) 176.2 | ||
| + | |||
| + | K243004 (a) 122.3, (b) 110.3 | ||
| + | |||
| + | I711009 (a) 185.2, (b) 174.7 | ||
| + | |||
| + | K157012 (a) 181.2, (b) 195.3 | ||
| + | |||
| + | K404108 (a) 116.4, (b) 142.5 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MidiPrep of pSB1C3 with RFP generator (P4) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of pSB1C3 with RFP-Reportercasette for further processing (P74) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of IRES-BirA (F33->F40) into digested F31 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: JH, LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of IRES-BirA (F29->F33->F40) into F31 (digested P40 by XbaI and PstI = pSB1C3 without insert), gelelectrophoresis and tranformation in XL1blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * F33 (PCR product of F29/gen synthesis 2) was digested with XbaI and PstI | ||
| + | * Gelextraction of band at 1500 bp was executed by manufacturers protocol (Qiagen) | ||
| + | * intermediate result: F40 with 6.6 ng/µL | ||
| + | * Ligation mixtures were prepared as following: | ||
| + | * 9.6 µL F40, 0.4 µL F31, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * Transformation: (Saturday) | ||
| + | * Transformation of Ligation (F29+F31) into XL-1 blue was done according to the SOP. The plate was incubated until Sunday at 37°C. | ||
| + | * 3mL-cultures in LB+ 30µg/ml cam where inoculated on Sunday | ||
| + | * Mini-prep according to manufacturer's protocol was done on Monday. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR, purification and digestion of P19 (EspP) and F28 (mRuby EGFR) for BioBrick creation === | ||
| + | |||
| + | '''Investigator: JB, LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of EspP from P19 and mRuby/EGFR (F28) for creation of Biobricks | ||
| + | |||
| + | '''Procedure (for both fragments):''' | ||
| + | * PCR according to the SOP (primers for P19: O13 and O03, primers for F28: VR and VF2) | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol (concentrations: P19(EspP):119,5 ng/µl F28(mRuby):56,2ng/µl | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | * Restriction digestion with NgoMIV/SpeI (for both cases) | ||
| + | * 40µl DNA | ||
| + | * 1,5µl of each Enzyme | ||
| + | * 2µl ddH2O | ||
| + | * 5µl CutSmart Buffer | ||
| + | * The tubes where incubated at 37°C for ~2 days. | ||
| + | |||
| + | '''Results:''' | ||
| + | Analytical gel electrophoresis after PCR purification shows thick bands at roughly 1000 bp | ||
| + | |||
| + | '''Still to do:''' | ||
| + | * Gel extraction of both signals for F28 and one signal for EspP-PCR | ||
| + | * Ligation with appropriate vector (e.g. F30) | ||
| + | * Transformation | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Minipreps of F39 (strong RBS-pcat in pSB1C3) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of F39 (ligation, pCat with strong RBS) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Miniprep according to the manufacturer's protocol | ||
| + | * Resulting plasmid: P76 | ||
| + | * Concentration: 52 ng/µl | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Minipreps of pSB1C3 with RFP generator (P4) === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of pSB1C3 with RFP-Reportercasette for further processing (P75) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Minipreps according to the manufacturer's protocol (with 8* 6 mL culture) | ||
| + | * Resulting plasmid: P75 | ||
| + | * Concentration: 142.2 ng/µl [~200 µL] | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of colony from NanoLuc [P54, Repeat] === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 ml LB+Cam media | ||
| + | * culture was inoculated with one colony | ||
| + | * Incubation at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, June 18th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Miniprep of P54 (NanoLuc) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of P54 (NanoLuc) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Miniprep according to the manufacturer's protocol | ||
| + | * Resulting plasmid: pSB1C3 mit NanoLuc (P54, von ) | ||
| + | * Concentration: 497,9 ng/µl | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Hybridisation of O19 and O20 (StrepTag) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Hybridisation of the Strep-Oligos | ||
| + | |||
| + | '''Procedure:''' | ||
| + | 2µl of each Oligo where diluted in 16µl ddH2O and incubated for 10 min. at 95°. Cooling down at RT for at least half an hour. Stored in fridge | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 6 (June 20th - June 26th)= | ||
| + | <div class="week" id="Week_6"> | ||
| + | |||
| + | =='''Monday, June 20th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Miniprep of F42 (IRES-BirA in pSB1C3) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of IRES-BirA Plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Miniprep according to the manufacturer's protocol | ||
| + | * Resulting plasmid: pSB1C3 with IRES-BirA | ||
| + | Next Steps: | ||
| + | * analytical digestion with EcoRI+PstI and gel (expected bands at 1500 and 2000 Bp) | ||
| + | * Sequencing | ||
| + | |||
| + | '''Results:''' | ||
| + | Concentration: 583,5 ng/µl | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR and Purification of BirA === | ||
| + | |||
| + | '''Investigator: LK, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of BirA-construct | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR according to the SOP (primers: O17 and O18) | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | |||
| + | '''Results:''' | ||
| + | * Concentration: 185,8 ng/µl --> F41 | ||
| + | * Analytical gel shows no significant band at 960 Bp -->Repeat | ||
| + | Next Steps: | ||
| + | * Repetition of PCR and analytical gel! | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Gel and Gelextraction of P19 (QC EspP) and F28 (EspP and mRuby) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purification of both constructs for Ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Preperative gel according to SOP | ||
| + | * The Purification was done according to the manufacturer's protocol. The EGFR gel-fragment was eluted in 50µl and the mRuby-fragment in 30µl (because of thin band) | ||
| + | |||
| + | '''Results:''' | ||
| + | * One thick band just before 1000Bp and one lighter band just before 750 Bp. This fits to the lengths of EspP and mRuby. | ||
| + | * Concentrations: EspP(now F43): 49,5ng/µl mRuby(now F44): 9,9ng/µl | ||
| + | |||
| + | Next Steps: | ||
| + | |||
| + | * Ligation into pSB1C3 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Sequencing of F42 (pSB1C3(IRES-BirA)), P54 (pSB1C3(NanoLuc)) , P74 (pSB1C3(RFP generator)) and P76 (pSB1C3(strong RBS-pCat))=== | ||
| + | |||
| + | '''Investigator: JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of F42, P54, P74 and P76 in forward direction. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | |||
| + | The different plasmids we prepared received the following barcodes: | ||
| + | * F42 (VF2): FR11326601 | ||
| + | * P54 (VF2): FR11326600 | ||
| + | * P74 (VF2): FR11326599 | ||
| + | * P76 (VF2): FR11326598 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === PCR and Purification of genesynthesis 6 (iGEM6) Streptavidin/Traptactin === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of gen synthesis 6 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | PCR according to the SOP (primers: VF2 and VR) | ||
| + | |||
| + | Next Steps: | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR and Purification of BirA (F41) === | ||
| + | |||
| + | '''Investigator: NA ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of BirA (F29) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | PCR according to the SOP (primers: O17 and O18) | ||
| + | |||
| + | Next Steps: | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * Analysis on an 1% agarose gel using 5 µl purified PCR product | ||
| + | |||
| + | --> repeat with gene-synthesis 2 (F29 may not be the gene-synthesis 2) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Ligations of StrepTag, EspP and mRuby3 with F30 (digested pSB1C3) === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligations of StrepTag (O19+O20), EspP (F43) and mRuby3 (F44) into F30 (NgoMIV and SpeI; pSB1C3 without insert) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ligation mixtures were prepared as following: | ||
| + | * O19+O20: 1.7 (O19+O20), 8.3 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * F43: 4.6 μL F43, 5.4 µL F30, 2 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * F44: 8.1 μL F44, 1.9 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * Ligation was incubated overnight at 16 °C | ||
| + | </div> | ||
| + | <div class="streptavidin"> | ||
| + | === Digestion of GSY5 (Streptactin; F27)=== | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Restricting GSY5 into StrepActin fragment (Streptavidin mutant) and Leptin fragment (Leptin is for Volker) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Restriction mix: | ||
| + | |||
| + | 32µl DNA (F27) | ||
| + | |||
| + | 10µl ddH2O | ||
| + | |||
| + | 1µl of each enzyme (SpeI, HindIII, SapI) | ||
| + | |||
| + | 5µl cut smart buffer | ||
| + | |||
| + | Next Steps: | ||
| + | * Preperative gel, 1,5% Agarose, cut out and purify bands at 80 Bp and 425 Bp. (80 is leptin for volker and 425 is StreptActin for further cloning into pASK) | ||
| + | |||
| + | '''Results:''' | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 21st'''== | ||
| + | <div class="general"> | ||
| + | === Plating of BioBricks that didn't survive sequencing === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Instead of directly inoculating cultures with the transformed cells from the bacterial agar that was sent by the HQ, the bacterial agar was now used for overnight plating on agar plates containing the appropriate antibiotic resistance. | ||
| + | |||
| + | '''Procedure:''' Two methods were used for each BioBrick: For the first method, an inoculation loop was used for plating directly. For the second methods, the inoculation loop was used to inoculate a 50 µl LB culture that | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | ===Sequencing of various plasmids === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of various plasmids with either VF2 or VR | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Refer to the “Inventarliste” for detailed informations on sequencing codes and primers. Sequencing batches were prepared according to manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR of gene synthesises (iGEM_1) (mRuby_EGFR) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' iGEM_1_mRuby_EGFR (F28) for preparing the EGFR signal (at about 100 bp) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Approach: | ||
| + | * 1µl DNA (diluted, 1:10) | ||
| + | * 25 µl 2x MasterMix | ||
| + | * 0,5µl dNTPs | ||
| + | * 2,5 µl forward Primer VF2 | ||
| + | *2,5µl reverse Primer VR | ||
| + | * Fill up with water up to 50 µl | ||
| + | Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/) | ||
| + | * 98°C_1min initiation step | ||
| + | * 98°C_10sec | ||
| + | * 66°C_30sec > step 2-4: 35 repeats | ||
| + | * 72°C_30sec | ||
| + | * 72°C_2min final step | ||
| + | * 4°_hold | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Transformation of Ligations of F50 (StrepTag), F51 (EspP) and F52 (mRuby3) in pSB1C3 === | ||
| + | |||
| + | '''Investigator: JH''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of StrepTag (F50), EspP (F51) and mRuby3 (F52) in XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. Coli XL1 blue according to protocol + rescue | ||
| + | (repeat due to incubation with LB+Cam ! ) | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Digestion of P9 (pASK75), P74 (pSB1C3), P26 (CMV promotor) and F47 (BirA) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of P9 and P74 to gain the vectors pASK75 and pSB1C3, respectively, as well as digestion of F47 (PCR fragment of BirA) and P26 (CMV promoter) for the ultimate creation of biobricks | ||
| + | |||
| + | '''Procedure:''' P74 was digested in a total volume of 30 µl, while the others were digested in a total volume of 50 µl. Enzymes used were: | ||
| + | * P74: XbaI and PstI | ||
| + | * P9: HindIII and XbaI | ||
| + | * F47: Xba I and PstI (didn’t work) | ||
| + | * P26: SpeI and PstI | ||
| + | The digestions were incubated at 37°C for 40 minutes and were thereafter inactivated at 80°C for 5 mins. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of cultures for minipreps for P76 (pCat + strong RBS) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Procedure:''' Inoculation of 2x5 ml cultures with P76 clones. Incubation at 37°C overnight. | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 22nd'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Minipreps of P79 (pCat+RBS) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Procedure:''' According to the manufacturer's protocol. | ||
| + | |||
| + | '''Results:''' Concentration 212 ng/µl. Sent to sequencing | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Transformation of ligation F56 (pASK75(Streptactin)), P60 (pSB1C3 (pC+AraC+pBad+GFPc)) , P70 (pSB1C3 (short linker)) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | ''Aim of the experiment:''' Transformation of pASK75-Streptactin (F56) into E. ''coli'' XL1 blue, K577881 (P60) into BL21 and K243004 (P70) in XL1 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. Coli XL1 blue according to protocol | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB + appropriate antibiotic) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of colonies from K157001 (EGFR signal peptide), EspP (F50), mRuby (F51), Strep-Tag (F52) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2x 5 ml LB+Cam (Amp for K157001) media | ||
| + | * Each culture was inoculated with one colony, for ligations 2 colonies each | ||
| + | * Incubation at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Plateing of K243004 (short linker), K243005 (middle linker) from stab cultures === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | For both biobricks: stabing into the agar with the inocluation loop and resuspending in 50 µl LB media. Plating on Amp-agar. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Digestion of P64 (EGFR sgnal peptide) and P71 (CD4 signal peptide) for cloning into F57 (pSB1C3 (CMV promotor)) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of P67 and P71 for cloning the resulting fragments into the digested F57 fragement. | ||
| + | |||
| + | '''Procedure:''' both plasmids were digested in 50 µl, 3 µg for each plasmid DNA. 1 µl of XbaI and PstI, 5 µl Buffer, 20 µl DNA, 23 µl H2O. The digestions were incubated at 37°C overnight. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Preparative agarose gels for separation of digestion of P64 (EGFR sgnal peptide) and P71 (CD4 signal peptide) === | ||
| + | |||
| + | '''Investigators: NA, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Separation of digestion fragments of both digested PCR fragments as well as vectors | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * P74: XbaI and PstI -> Fragment corresponding to the pSB1C3 vector isolated | ||
| + | * P9: HindIII and XbaI -> Fragment corresponding to the pASK75 vector isolated | ||
| + | * F47: Xba I and PstI (didn’t work) –> only one fragment at 3 kbp present (weird since that was a gene synthesis pcr) | ||
| + | * P26: SpeI and PstI -> Fragment corresponding to the cmv-promoter isolated | ||
| + | |||
| + | [[File:Muc16_praep_p74_p9.png|500px]] | ||
| + | |||
| + | [[File:Muc16_praep_p26.png|500px]] | ||
| + | |||
| + | * Gelextraction of P9, P74 and P26 was executed by manufacturers protocol (Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * F55: pASK75 (2,9 ng/µl) | ||
| + | * F56: pSB1C3 (23,6 ng/µl) | ||
| + | * F57: CMV-Promotor (5,9 ng/µl) | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR of GSY2 (IRES-BirA) with O17/18 === | ||
| + | |||
| + | '''Investigators: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of BirA, using new dilution of GSY2 this time, because last 2 PCRs showed confusing results (bands at 2500) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR was performed according SOP | ||
| + | * Primers: O17 and O18 | ||
| + | * Annealing at 68°C | ||
| + | |||
| + | Next Steps: | ||
| + | * PCR-Purification | ||
| + | * Analytical Gel | ||
| + | * Ligation into pSB1C3 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === QuickChange-PCR of pASK75 with O21 and O22 === | ||
| + | |||
| + | '''Investigators: LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Deleting NdeI from pASK (P03) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * QC-PCR was performed according to SOP | ||
| + | * Primers: O-021 and O-022 | ||
| + | * Annealing at 68°C | ||
| + | * Elongation time: 3:30 min | ||
| + | |||
| + | Next Steps: | ||
| + | * Digestion with DpnI at 37°C for 1h | ||
| + | * Purification? | ||
| + | * Preperative Gel (band at 960), purification | ||
| + | * Ligation into pSB1C3 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Verification of PCR and Digestion of F28 (gene synthesis 1) === | ||
| + | |||
| + | '''Investigator: NA, LK''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful PCR via gel electrophoresis and preparation of EGFR | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Gel electrophoresis with 1 kb-ladder | ||
| + | * Digestion: 42µl of PCR Purification, 1,5 µl of each enzyme (NgoMIV and SpeI), 5µl of cut smart buffer. | ||
| + | * Incubation at 37°C over night | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_analytic_after_pcr_F28.png|500px]] | ||
| + | |||
| + | Next steps: | ||
| + | * Preparative gel and gel extraction | ||
| + | * Ligation into pSB1C3 | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, June 23th'''== | ||
| + | <div class="receptor"> | ||
===Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001=== | ===Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001=== | ||
| Line 421: | Line 2,969: | ||
'''Aim of the experiment:''' Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001 | '''Aim of the experiment:''' Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001 | ||
| + | |||
'''Procedure:''' | '''Procedure:''' | ||
* Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
* Concentrations: | * Concentrations: | ||
| − | <table cellspacing="0" border="1"> | + | <table class="wikitable" cellspacing="0" border="1"> |
<tr> | <tr> | ||
<td><b>Plasmid</b> | <td><b>Plasmid</b> | ||
| Line 459: | Line 3,008: | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="receptor"> |
| + | === PCR of gene synthesises === | ||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' iGEM_1_mRuby_EGFR (F28) for preparing the EGFR signal (at about 100 bp) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Approach: | ||
| + | * 1µl DNA (diluted, 1:10) | ||
| + | * 25 µl 2x MasterMix | ||
| + | * 0,5µl dNTPs | ||
| + | * 2,5 µl forward Primer VF2 | ||
| + | * 2,5µl reverse Primer VR | ||
| + | * Fill up with water up to 50 µl | ||
| + | |||
| + | Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/) | ||
| + | * 98°C_1min initiation step | ||
| + | * 98°C_10sec | ||
| + | * 66°C_30sec > step 2-4: 35 repeats | ||
| + | * 72°C_30sec | ||
| + | * 72°C_2min final step | ||
| + | * 4°_hold | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === QuickChange-PCR of pASK with O21 and O22 === | ||
| + | |||
| + | '''Investigators: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of PCR product with DpnI to digest methylated DNA | ||
| + | |||
| + | '''Procedure:''' | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === PCR purification of BirA and mRuby/EGFR === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of PCR | ||
| + | |||
| + | '''Procedure:''' | ||
| + | PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | The samples were renamed: | ||
| + | * BirA: F59 | ||
| + | * mRuby/EGFP: F60 | ||
| + | The samples were analysed on a 1% agarose gel. Samples were prepared after SOP. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_pcr_F59_F60.png|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Transformation of P60 (K577881; pC+AraC+pBad+GFPc K1) and QC P3 (pASK75(SA1)) === | ||
| + | |||
| + | '''Investigator: EF, JB''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of Q CP3 in XL1 blue; transformation of p60 in BL21 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. Coli BL21 and XL1blue according to protocol | ||
| + | |||
| + | 1µl p60 in BL21 | ||
| + | 7 µl Q CP3 in XL1 blue | ||
| + | |||
| + | '''Result:''' plates (+ rescue plates) incubating | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of CD4- and EGFR-signal peptide into pSB1C3-CMV === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion of P71 and P69 | ||
| + | '''Procedure:''' | ||
| + | * Digestions were prepared as following: | ||
| + | * 20 µl CD4 (P71); 1 µl XbaI; 1 µl PstI; 5 µl CutSmart; 23 µl ddH2O | ||
| + | * 7 µl pSB1C3-CMV; P26; 1 µl PstI; 1 µl SpeI; 2 µl CutSmart; µl ddH2O | ||
| + | * 20 µl EGFR-signal peptide (P67);1 µl XbaI; 1 µl PstI; 5 µl CutSmart; 23 µl ddH2O | ||
| + | * Digestions were incubated for 3 h at 37 °C | ||
| + | * The Samples CD4- and EGFR-signal peptide were applied to a 2% and the pSB1C3-CMV to a 1% agarose Gel | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Cloning_of_CD4-and_EGFR-signal_peptide_into_pSB1C3-CMV_1.png|500px]] | ||
| + | |||
| + | --> passt | ||
| + | |||
| + | [[File:Muc16_Cloning_of_CD4-and_EGFR-signal_peptide_into_pSB1C3-CMV_2.png|500px]] | ||
| + | |||
| + | --> passt | ||
| + | |||
| + | Next steps: | ||
| + | * Ligation of | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of EGFR (F62) / CD4 (F61) signale peptide into pSB1C3-CMV (F63) === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Gelextraction and ligation of EGFR- and CD4-signal peptide into pSB1C3 and transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Gel extraction according to manufacturers protocol (Qiagen): F61 (CD4) 2 ng/µl, F62 (EGFR) 0.6 ng/ µl, F63 (CMV): 45 ng/ µl | ||
| + | * Ligation: 1 µl T4 Ligase, 2 µl Ligase buffer, 7 µl H20, 1.8 µl vector and 8.2 µl EGFR insert or 4.3 µl vector and 5.7 µl CD4 insert | ||
| + | * Transformation according to SOP | ||
| + | * Plating on LB-Cam agar | ||
| + | </div> | ||
=='''Friday, June 24th'''== | =='''Friday, June 24th'''== | ||
| − | <div class=" | + | <div class="receptor"> |
=== Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2) === | === Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2) === | ||
| Line 496: | Line 3,156: | ||
[[File:Muc16_P82-85_EcoRI.png|500px]] | [[File:Muc16_P82-85_EcoRI.png|500px]] | ||
| + | [[File:Muc16_P82-85_EcoR.png|500px]] | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="receptor"> |
| − | === Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)=== | + | === Sequencing of P80( mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)=== |
'''Investigator: Julian''' | '''Investigator: Julian''' | ||
| Line 520: | Line 3,181: | ||
</div> | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of ''E. coli'' XL1 blue with F64 (quickchanged P3(pSAm1)) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 1 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Gelextraction of F67(BirA(Digest. F59 [EcoRI; SpeI])), F68(mRuby(Digest. F60 [NgoMIV; SpeI])), F69(EGFR-TMD(Digest. F60 [NgoMIV; SpeI]), F70(pSB1C3(digest. P74 [NgoMIV; SpeI]) and F71(pSB1C3(digest. P74 [EcoRI; SpeI]) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Gelextraction was performed by manufacturers protocol (Qiagen). | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of F59 (BirA) and F60 (EGFR-TMD) into pSB1C3 === | ||
| + | |||
| + | '''Investigator: CR''' | ||
| + | |||
| + | '''Aim of the experiment:''' Generate biobricks with EGFR-TMD and BirA | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestions were prepared as following: | ||
| + | * 1. xµl F59 (BirA); 1 µl EcoRI; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O | ||
| + | * 2. 2 µl P74 (pSB1C3 (RFP-generator); 1 µl NgoMIV; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O | ||
| + | * 3. xµl F60 (mRuby/EGFR); 1 µl NgoMIV; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O | ||
| + | * 4. 2 µl P74 (pSB1C3 (RFP-generator), 1 µl EcoRI; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O | ||
| + | * Digestion was incubated for 3 h at 37 °C | ||
| + | |||
| + | To Do: | ||
| + | * Gel (already prepared) | ||
| + | * Gelextraction | ||
| + | * Ligation | ||
| + | * Trafo | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Cloning_of_BirA_and_EGFR-TMD_into_pSB1C3.png|500px]] | ||
| + | |||
| + | 1. band: P74(NgoMiV, SpeI) 2. band: P74 (EcoRI, SpeI) | ||
| + | |||
| + | [[File:Muc16_Cloning_of_BirA_and_EGFR-TMD_into_pSB1C3_2.png|500px]] | ||
| + | |||
| + | 1. band: F59 2. band: F60 | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Transformation of EGFR-TMD-pSB1C3 and BirA-pSB1C3 ligations === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Procedure:''' According to protocol. Plates were incubated overnight at 37°C. | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Chemical biotinylation of BSA === | ||
| + | |||
| + | '''Investigator: MT, JB''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | BSA was chemically biotinylated with a 20x, 40x and 60x molar excess in two different buffers:100 mM Bicin (pH 8.85) and 100 mM borate (pH 8.85) with 50 mM NaCl respectively. | ||
| + | |||
| + | -> A total of six different tubes. | ||
| + | |||
| + | After one hour incubation at RT, the samples were put in the freezer | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Inoculation of liquid cultures (F65 and F66) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Clones were picked for inoculation of 5 ml liquid cultures containing the appropriate antibiotic | ||
| + | * Incubation overnight at 37°C/shaking | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, June 25th'''== | ||
| + | <div class="receptor"> | ||
| + | ===Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P87 | ||
| + | </td><td>81 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P88 | ||
| + | </td><td>34,5 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P89 | ||
| + | </td><td>86,3 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P90 | ||
| + | </td><td>108,5 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P91 | ||
| + | </td><td>417,4 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of F64 (quickchanged P3, pASK75(SA1)) for verification of success of quickchange === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Analytical digestion with NdeI and gelelectrophoresis. If quickchange worked there should be a band at about 3200 bp (only one restriction site left) | ||
| + | |||
| + | * Incubation over night at room temperature. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_Quickchange_NA.JPG]] | ||
| + | |||
| + | Just a band showing a few bp (Primer), there is no plasmid band -> Quickchange did not work | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3) and F44 + F33 (mRuby in pSB1C3) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 6x 4 ml LB+Cam media | ||
| + | |||
| + | * Each culture was inoculated with one colony | ||
| + | |||
| + | * Incubation at 37°C overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Sunday, June 26th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extracting F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) from E.coli XL-1-blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P92 | ||
| + | </td><td>162,9 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P93 | ||
| + | </td><td>447,9 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Repetition of Quick-Change PCR of P3 (pASK + SAm1) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The QC-PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O21 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O22 | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 5 µl | ||
| + | </td><td>Pfu-Ultra-II reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td>Pfu-Ultra-II Polymerase | ||
| + | <tr> | ||
| + | <td>40,5 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | * digestion of PCR-Product with DpnI for 1h at 37°C | ||
| + | |||
| + | * now labeled P94 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Transformation of ''E. coli'' XL1 blue with P94 (quickchanged P3(pSAm1)) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) for pASK (F72) and on chloramphenicol plates for P92 and P93 and incubated over night at 37 °C in the incubator. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75). | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P92 and P93 with EcoRI-HF and PstI-HF | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl) and PstI-HF (10 U/µl) for P92/93, NdeI (10 U/µl) for P94 | ||
| + | |- | ||
| + | |7.5/7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * 1. band (P92): no mRuby at 700bp visible, only empty vector | ||
| + | * 2. band (P93): empty vector and EGFR --> perfect | ||
| + | * 3. band (P94): no signal at all --> repetition of QC-PCR | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
<!--- this closes the week --> | <!--- this closes the week --> | ||
| Line 526: | Line 3,497: | ||
<!--- PLEASE DO NOT TOUCH !!!! --> | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
<!--- PLEASE DO NOT TOUCH !!!! --> | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 7 (June 27th - July 3rd)= | ||
| + | <div class="week" id="Week_7"> | ||
| + | |||
| + | =='''Monday, June 27th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Sequencing of P67 (EGFR-Signalpeptid) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2)) | ||
| + | |||
| + | FR11326586 | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Repetition of Quick-Change PCR of P3 (pASK + SAm1) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The QC-PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O21 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>1,25 µl | ||
| + | </td><td>Primer O22 | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 5 µl | ||
| + | </td><td>Pfu-Ultra-II reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td>Pfu-Ultra-II Polymerase | ||
| + | <tr> | ||
| + | <td>40,5 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | * Digestion of PCR-Product with DpnI for 1h at 37°C. | ||
| + | |||
| + | * Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR). | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === PCR of Genesynthesis 3 and 4 === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * The PCR was performed according the SOP. | ||
| + | |||
| + | * Reaction Mix: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>volume</b> | ||
| + | </td><td><b>reagent</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 2,5 µl | ||
| + | </td><td>Primer VF2 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 2,5 µl | ||
| + | </td><td>Primer VR2 | ||
| + | <tr> | ||
| + | <td> 1 µl | ||
| + | </td><td> dNTP-mix | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 10 µl | ||
| + | </td><td> Q5 Polymerase reaction buffer | ||
| + | <tr> | ||
| + | <td>1 µl | ||
| + | </td><td> template DNA (1:10 dilution of p3) | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 0,5 µl | ||
| + | </td><td> Q5-Polymerase | ||
| + | <tr> | ||
| + | <td> 18 µl | ||
| + | </td><td> ddH2O | ||
| + | </td></tr></table> | ||
| + | |||
| + | *Setup: iGEM_standard (Promega-cycler) | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>temperature</b> | ||
| + | </td><td><b>time</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 98°C | ||
| + | </td><td> 2min | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 98°C | ||
| + | </td><td> 10sec | ||
| + | <tr> | ||
| + | <td> 66°C | ||
| + | </td><td> 30sec | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 72°C | ||
| + | </td><td> 30sec | ||
| + | <tr> | ||
| + | <td> 72°C | ||
| + | </td><td> 2min | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td> 4°C | ||
| + | </td><td> hold | ||
| + | </td></tr></table> | ||
| + | |||
| + | * the batches were then purified using the Quiagen PCR-Purification Kit. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Analytical digestion and gelelectrophoresis of P88 (pASK75(Steptactin)), P89 (pSB1C3(CMV+CD4) and P90 (pSB1C3(CMV+EGFR-Signal peptide) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | P88: EcoRI | ||
| + | |||
| + | P89: EcoRI and PstI | ||
| + | |||
| + | P90: EcoRI | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |5,8/2,3/1,8 µl | ||
| + | |Plasmid DNA (P88/P89/P90) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl)/ PstI | ||
| + | |- | ||
| + | |required amount for total volume of 10 µl | ||
| + | |ddH2O | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_P88-90_NA.JPG]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Ligation of F67 (BirA) and F71 (vector fragment pSB1C3), Transformation of ''E. coli'' XL1 blue afterwards === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of ''E. coli'' XL1 blue afterwards. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2,4 µl | ||
| + | |Vektor | ||
| + | |- | ||
| + | |7,6 µl | ||
| + | |Insert | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | * next step: analytic digestion of transformation was successful | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Digestion of PCR on genesynthesis 3 and 4, and pSB1C3 === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl each | ||
| + | |enzyme (SapI, HindIII, XbaI, AgeI) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |41 µl | ||
| + | |DNA (purified PCR-products of GSY3 and 4) | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | * Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Preparative digestion and gelelectrophoresis of P80 , P78 and P85 === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: Preparative digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | *P80 and P78: EcoRI and AgeI | ||
| + | |||
| + | *P85: EcoRI and NgoMIV | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |32 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |each enzyme(10 U/µl)/ PstI | ||
| + | |- | ||
| + | |50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_P78-P80_2806.jpeg]]<br> | ||
| + | [[File:Muc16_P85_2806.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into ''E. coli'' XL1 blue === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of ''E. coli'' XL1 blue afterwards. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |4,3µl(for F75), 8,2µl (for F76) | ||
| + | |Vector | ||
| + | |- | ||
| + | |12,7µl (F75), 8,8 (F76) | ||
| + | |Insert | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Ligation was incubated at RT for 1,5h. | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 15 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Chemical biotinylation of BSA === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * BSA was chemically biotinylated with a 20x and 40x molar excess: | ||
| + | |||
| + | * 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85) | ||
| + | |||
| + | * dissolve BSA (10 mg/ml) | ||
| + | |||
| + | * Add biotin-NHS-ester: 20,5 mg for 40x molar excess | ||
| + | |||
| + | * reaction over night | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, June 28th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | *P83: EcoRI and SpeI | ||
| + | |||
| + | *P29: EcoRI and XbaI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6.5/13 µl | ||
| + | |Plasmid DNA (P83/P29) | ||
| + | |- | ||
| + | |35.5/29 µl | ||
| + | |ddH2O (P83/P29) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |each enzyme | ||
| + | |- | ||
| + | |50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | *Preparative gelelectrophoresis was performed at 80 V for 80 min | ||
| + | |||
| + | [[File:Muc16_P83P29_2806.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Gelelectrophoresis of F73 (pSB1C3 digst. EcoRI, AgeI) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Preparative gelelectrophoresis was performed at 100 V for 45 min using a pocket spanning over the full width of the 1% agarose gel | ||
| + | |||
| + | [[File:Muc16_F71_2806.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, June 29th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Transformation of F83 (EspP-Fragment in Double Terminator + pSB1C3), F84 (GSY3 + pSB1C3) F85 (GSY4 + pSB1C3) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, June 30th'''== | ||
| + | |||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Transformation of new ''E. coli'' XL1-blue (P75), additional plating of untransformated XL1-blue === | ||
| + | |||
| + | '''Aim of experiment: Quality assurance''' | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the new competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 1 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C. | ||
| + | |||
| + | * additional plating of untransformated XL1-blue on Tet-, Cam-, Amp- and Kan-plates | ||
| + | |||
| + | '''Result:''' | ||
| + | |||
| + | * After 12 hous of incubation there were some (~20) colonies on the 50µl-plate transformed with Cam-resistence-plasmids. The transformation has been succesful, though cells grow slow. | ||
| + | * There were no colonies on the kan-, amp- and cam- plates with the untransformed E.Coli. Hence we can assume that there are no other plasmids with resistences against kan, amp or cam in our batch of competent cells. | ||
| + | |||
| + | There are cells on the tet-plate. This was expected. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, June 31th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of P9 (pASK with Streptavidin WT) and QC-PCR product into new ''E. coli'' XL1-blue === | ||
| + | |||
| + | '''Aim of experiment: Tranforming pASK without NdeI-cutting point into E.coli (two batches from diffrent approches were tested). And Transforming pASK_SA1 into E.coli XL-1-blue.''' | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the new competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 15 min incubation on ice | ||
| + | |||
| + | * 30 sec. heat shock at 42 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. 1h hour incubation at 37°C. | ||
| + | |||
| + | * The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | =Week 8 (July 04–10)= | ||
| + | <div class="week" id="Week_8"> | ||
| + | |||
| + | =='''Monday, July 04'''== | ||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Repetition of the Transformation of QC-PCR product into new ''E. coli'' XL1-blue === | ||
| + | |||
| + | '''Aim of experiment: Tranforming pASK without NdeI-cutting point into E.coli (two batches from diffrent approches were tested).''' | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the new competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 15 min incubation on ice | ||
| + | |||
| + | * 30 sec. heat shock at 42 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. 1h hour incubation at 37°C. | ||
| + | |||
| + | * The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of P9 (Streptavidin in pASK75) into ''E. coli'' BL-21 === | ||
| + | |||
| + | '''Aim of experiment: Tranforming pASK with Streptavidin into E.coli BL-21 for Expression.''' | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the new competent ''E. coli'' BL-21 cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 1 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 15 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of F94 (hGH polyadenylation sequence and Nanoluc with StrepTag) === | ||
| + | |||
| + | '''Investigator: Javier''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 45 sec. heat shock at 42 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium. | ||
| + | |||
| + | * The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | ===Miniprep of E. coli Xl1-Blue transformed with ligation product P108 and P113 (EspP-Fragment in Double Terminator, F81+F82), P109 and P110 (GSY3+ F73 vector AgeI+XbaI ), P111 and P112 (GSY4+ F73 vector AgeI+XbaI) === | ||
| + | |||
| + | '''Investigator: Javier''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extracting P108 and P113 (EspP-Fragment in Double Terminator, F81+F82), P109 and P110 (GSY3+ F73 vector AgeI+XbaI ), P111 and P112 (GSY4+ F73 vector AgeI+XbaI) from E.coli XL-1-blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P108 | ||
| + | </td><td>116,2 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P109 | ||
| + | </td><td>86,6 | ||
| + | <tr> | ||
| + | <td>P110 | ||
| + | </td><td>136,8 | ||
| + | <tr> | ||
| + | <td>P111 | ||
| + | </td><td>144,8 | ||
| + | <tr> | ||
| + | <td>P112 | ||
| + | </td><td>163,2 | ||
| + | <tr> | ||
| + | <td>P113 | ||
| + | </td><td>123,3 | ||
| + | </td></tr></table> | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Preparative digestion and gelelectrophoresis of P74 (RFP-Gen.) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: Preparative digestion and gelelectrophoresis of P74 (RFP-Gen.) ''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion: | ||
| + | |||
| + | *P74: EcoRI and SpeI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |39.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |each enzyme | ||
| + | |- | ||
| + | |50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Preparative gelelectrophoresis was performed at 90 V for 45 min | ||
| + | |||
| + | '''Result: '''pSB1C3 (EcoRI und SpeI) F95 | ||
| + | |||
| + | [[File:Muc16_F95_0407.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of F96 (BirA in pSB1C3) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 10 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 5 min incubation on ice | ||
| + | |||
| + | * 30 s heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium | ||
| + | |||
| + | * Incubation at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, July 05th'''== | ||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Preparing of 200 ml TB medium for inoculation of 2 L medium on Thursday === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 2.4 g Tryptone 4.8 g yeast extract | ||
| + | |||
| + | * 800 µl Glycerine | ||
| + | |||
| + | * adjust to 180 ml with ELGA-water | ||
| + | |||
| + | * seperately: 3.29 g H<sub>2</sub>KPO<sub>4</sub> + 0,46 g K<sub>2</sub>HPO<sub>4</sub> with ELGA water (Volume = 20 ml) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Digestion of F49 (BirA) with EcoRI and SpeI and cloning in F95 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Digestion of F49 with EcoRI and SpeI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |20 µl | ||
| + | |Plasmid DNA (F49) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF(10 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |SpeI(10 U/µl) | ||
| + | |- | ||
| + | |23 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Digestion was incubated for 1.5 h at 37°C | ||
| + | *Digestion was applied to a 1% agarose-gel | ||
| + | *Gel ran at 80V for 35 min | ||
| + | |||
| + | [[File:Muc16_digestF49_0507.jpeg]] | ||
| + | |||
| + | *BirA had a length of 1012 bp | ||
| + | *Upper probe was cut from the gel | ||
| + | *BirA was purified with gelextraction which was performed by manufacturer's protocol | ||
| + | *Concentration was measured | ||
| + | **[F97]= 12.4 | ||
| + | *Ligation of F97 into pSB1C3 (F95) | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.2 µl | ||
| + | |Vector: pSB1C3 (F95) | ||
| + | |- | ||
| + | |8.8 µl | ||
| + | |Insert: BirA (F97) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |T4 DNA Ligase buffer | ||
| + | |- | ||
| + | |fill up to 20 µl | ||
| + | |dd H<sub>2</sub>O | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4 ligase | ||
| + | |- | ||
| + | |=21 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Ligation was incubated at RT for 30 min | ||
| + | *Transformation: | ||
| + | **Transformation was performed after SOP | ||
| + | **7 µl were used for transformation in E. ''coli'' XL1blue | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P108 - P112 === | ||
| + | |||
| + | '''Aim of experiment: Sequencing ''' | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * sequencing batches after protocol with following barcodes: | ||
| + | |||
| + | * P108 (EspP-Fragment in Double Terminator (F83)) FR11326572 (VF2) | ||
| + | |||
| + | * P109 (GSY3+ F73 Leervektor AgeI+XbaI (F84)) FR11326576 (VF2) | ||
| + | |||
| + | * P110 (GSY3+ F73 Leervektor AgeI+XbaI (F84)) FR11326575 (VF2) | ||
| + | |||
| + | * P111 (GSY4+ F73 Leervektor AgeI+XbaI (F84)) FR11326574 (VF2) | ||
| + | |||
| + | * P112 (GSY4+ F73 Leervektor AgeI+XbaI (F84)) FR11326573 (VF2) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === NEB Turbo CaCl<sub>2</sub>-competent cells === | ||
| + | |||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Inoculate 50 mL LB with 0.5 mL overnight culture of NEB Turbo cells | ||
| + | |||
| + | * Incubate at 37 °C for 7 h; final OD<sub>500</sub>: 0.357 | ||
| + | |||
| + | * Very little growth in the course of the last hour; OD<sub>500</sub> 0.45 required | ||
| + | |||
| + | * Experiment discontinued. Inoculated 3 further colonies in 5 mL LB for o/n growth (37 °C). Repeat experiment on Wed, July 6 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Miniprep P114 (NanoLuc + StrepTag + Poly-A in pSB1C3(F94)) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Analytical digestion and gelelectrophoresis of P114 and P115 === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Analytical digestion and gelelectrophoresis of P114 and P115 (NanoLuc + StrepTag + Poly-A in pSB1C3 K1 and K2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P114 and P115 with EcoRI+PstI | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.75 µl | ||
| + | |Plasmid DNA P114/P115 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |cut smart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |6,25 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | * Incubation for 45 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_digestP114_P115_0507.jpeg]] | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp ladder DNA ladder | ||
| + | |'''P114''' | ||
| + | |'''P115''' | ||
| + | |- | ||
| + | | | ||
| + | |'''X''' | ||
| + | |'''X''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inoculation of Quick change of P88 (pASK75 (Streptactin)) preculture === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * four colonies were picked from plate | ||
| + | * Cells were inoculated in 5 mL LB medium (ampicillin 1:1000) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion of P108 and P113 (both EspP + double terminator (F83)) with EoRI and PstI === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Digestion of F49 with EcoRI and SpeI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Plasmid DNA (P108/P113) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF(10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI(10 U/µl) | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Digestion was incubated for 1.5 h at 37°C | ||
| + | *Digestion was applied to a 1% agarose-gel | ||
| + | *Gel ran at 80V for 35 min | ||
| + | |||
| + | [[File:Muc16_analyt.digest.P108_P113_0507.jpeg]] | ||
| + | |||
| + | *P108 and P113 showed the right bands | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, July 06th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P108 (EspP-Fragment in Double Terminator (F83)) === | ||
| + | |||
| + | '''Investigator: >Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * sequencing batches after protocol with following barcodes: | ||
| + | |||
| + | * P108 (EspP-Fragment in Double Terminator (F83)) FR11326572 (VR) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Digestion of P75 (RFP-Generator) with HincII + dephosphorylation === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of experiment:''' Blunt-Ligation of BirA into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Digestion of P75 (RFP-Generator) with HincII + dephosphorylation | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Plasmid DNA (P75) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |HincII | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Fast AP (for dephosphorylation) | ||
| + | |- | ||
| + | |14.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Digestion was incubated for over night at 37°C | ||
| + | |||
| + | *next steps: PCR of GSY2 with O17 and O18 + phosphorylation, Ligation in digested pSB1C3 afterwards | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of A3C5 Tag into the MiddleLinker-Plasmid === | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim of experiment:''' Cloning of A3C5 Tag into the MiddleLinker-Plasmid | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Digestion of Middle_Linker Plasmid (P69) with AgeI and PstI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.3 µl | ||
| + | |Plasmid DNA (69) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Fast AP (for dephosphorylation) | ||
| + | |- | ||
| + | |14.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.3 µl | ||
| + | |Plasmid DNA (P112) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgomIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |14.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | *Digestion was incubated for over night at 37°C, heat inactivation at 80°C for 20 min. | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl | ||
| + | |digested Middle-Linker (F99) | ||
| + | |- | ||
| + | |8 µl | ||
| + | |digested A3C5 (F100) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10x Ligase Buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Transformation according to SOP in E. coli XL1 blue | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Picking of of ''E. coli'' XL1 blue with Streptactin in quickchanged pASK75, F84 (digested GSY3), F85 (digested GSY4) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Streptactin in quickchanged pASK75: Colonies were picked from Amp-plates. (10x) | ||
| + | |||
| + | * F84/F85: Colonies were picked from Cam-plates. (8x each) | ||
| + | |||
| + | * Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL of the proper antibiotic. | ||
| + | |||
| + | * These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Quick Change of mRuby Plasmids === | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim of experiment:''' Correction of wrong amino acid within the mRuby sequence | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | QC according to manufacturers protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 ng (= 0,3 µl 1:10) | ||
| + | |Plasmid DNA (80) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |10x Pfu Ultra II Buffer | ||
| + | |- | ||
| + | |1.25 µl | ||
| + | |O23 | ||
| + | |- | ||
| + | |1.25 µl | ||
| + | |O24 | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |Pfu Ultra II | ||
| + | |- | ||
| + | |1 µl | ||
| + | |dNTPs | ||
| + | |- | ||
| + | |40.7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 ng (= 0,3 µl 1:10) | ||
| + | |Plasmid DNA (100) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |10x Pfu Ultra II Buffer | ||
| + | |- | ||
| + | |1.25 µl | ||
| + | |O23 | ||
| + | |- | ||
| + | |1.25 µl | ||
| + | |O24 | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |Pfu Ultra II | ||
| + | |- | ||
| + | |1 µl | ||
| + | |dNTPs | ||
| + | |- | ||
| + | |40.7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *PCR: 2 min. 98°C initial denaturation, 10 sec. 98°C, 30 sec. Annealing st 60 °C, 5 min 68°C, 35 Cycles | ||
| + | |||
| + | *1 h at 37°C DpnI Digestion | ||
| + | |||
| + | *Transformation according to SOP in E. coli XL1 blue, QC on mRuby-Strep (F103) was also transformed in E. coli Turbo cells for further investigating the growth rate | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Miniprep of Quick Change of pASK75-Streptactin + analytical digestion with Nde1 === | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Miniprep using the kit according to the manufacturer's protocol. | ||
| + | |||
| + | |||
| + | *Although cultures seemed to have grown properly, very low and unreliable DNA concentrations were obtained via Nanodrop measurement and no fragment at all could be detected via analytical agarose gel electrophoresis after 90 minutes digestion with Nde1 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Creation of competent NEB Turbo cells=== | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 250 µl of precultures were used to inoculate 50 ml LB-medium | ||
| + | * After an OD=0.45 was reached, cells were put on ice and made competent according to the manufacturer's protocol | ||
| + | * 50 µl aliquots of competent cells were frozen in liquid nitrogen and stored at -80°C until further use | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | |||
| + | ===Inoculation of a BL21 pBAD-GFP (P60) preculture=== | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * A single clone was picked in order to inoculate 5 ml LB medium (+Cam) | ||
| + | * Culture was grown overnight at 37°C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Inoculation of a BL21 preculture for Streptavidin production (P8) === | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * A clone was picked for the inoculation of 5 ml LB medium | ||
| + | * After 6-8 hours of incubation, the 5 ml preculture was used for the inoculation of a 50 ml LB preculture | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, July 07'''== | ||
| + | <div class="general"> | ||
| + | ===PCR Amplification of ''birA'' (GSY 2)=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Amplify ''birA'' gene | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Prepared PCR mix (25 μL reaction volume): NEB Q5 High-Fidelity 2× Master Mix (12.5 μL), ddH<sub>2</sub>O (9.5 μL), F29 (1 μL), O17 and O18 (1 μL each) | ||
| + | |||
| + | * Cycling protocol: start 1.5 min, 98 °C; 35 cycles 30 s, 98 °C – 45 s, 72 °C – 30 s, 72 °C; final elongation 5 min, 72 °C; end 4 °C | ||
| + | |||
| + | * Purified ''via'' Quiagen PCR Purification kit following manufacturer's instructions. DNA eluted from column by adding 50 μL EB buffer and heating at 37 °C for 5 min prior to centrifugation. | ||
| + | |||
| + | '''Results:''' DNA concentration: 82.7 ng/μL. Stored as F102 at –20 °C | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | ===Induction of Streptavidin=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Induce streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Inoculated 2 L TB medium with 50 mL o/n pre-culture and added 2 mL ampicillin (10 mg/mL in 50% EtOH) | ||
| + | * Let grow (37 °C, 200 rpm) until OD<sub>500</sub>=0.622 (''c.'' 2.5 h) | ||
| + | * Induced with anhydrotetracycline (1:10,000 i.e. 200 μL of 2 mg/mL stock) | ||
| + | * Let grow (37 °C, 200 rpm) for 4 h | ||
| + | |||
| + | '''Results:''' Induced culture for harvesting (see below) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | ===Production of eGFP=== | ||
| + | '''Investigator: Clemens, Vivien, Christoph''' | ||
| + | |||
| + | '''Aim:''' Produce eGFP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Inoculated 50 mL LB+Cam medium with 0.5 mL o/n pre-culture | ||
| + | |||
| + | * Let grow (37 °C, 200 rpm) until OD<sub>500</sub>=0.58 | ||
| + | |||
| + | * Induced with 0.1 mM arabinose (i.e. 100 μL of 50 mM stock) | ||
| + | |||
| + | * After 7 h of induction the cells were stored in the fridge overnight for maturation of the GFP-chromophore | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Harvesting of Streptavidin=== | ||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor) | ||
| + | * The supernatant was discarded, and the pellet was transferred into a Falcon and frozen | ||
| + | * next steps: resuspending and homogenizing in the PANDA | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, July 08'''== | ||
| + | <div class="general"> | ||
| + | |||
| + | === Analytical Digest of mRuby3 Constructs === | ||
| + | |||
| + | '''Aim of experiment: Verify integration of mRuby3 constructs in F103 and F104''' | ||
| + | |||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Mixed 16 μL F103 (or 8 μL F104 plus 8 μL ddH<sub>2</sub>O) with 2 μL 10× CutSmart buffer and 1 μL each of EcoRI-HF and PstI-HF (20 μL total volume) | ||
| + | |||
| + | * Incubated for 45 min at 37 °C | ||
| + | |||
| + | * Subjected to gel electrophoresis (1% agarose, 80 V, 120 min) | ||
| + | |||
| + | '''Results:''' No band around 700 bp as expected for mRuby3 constructs | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Picking clones and inoculating cultures for F101 and F103 === | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * All clones available (only two for F101, one for F103) were picked and used to inoculate 5 ml of LB medium containing the appropriate antibiotic | ||
| + | * Cultures were incubated at 37°C overnight | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Solubilization, lysis and unfolding of streptavidin === | ||
| + | |||
| + | '''Investigator: Jan''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * According to protocol -> lysis of pellet (19.2 g) in 58 ml lysis buffer (50 mM Tris/HCl pH 8, 1 mM EDTA) | ||
| + | * As only two liters of medium were used, the Panda couldn't be utilized for cell lysis -> French Press was used | ||
| + | * Cells were additionally submitted to ultrasonic treatment | ||
| + | * Centrifugation of cells, casting away the supernatant and resolubilizing the pellet in 5 ml GdmCl (6M, pH 1.5) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Sequencing of P88 === | ||
| + | |||
| + | '''Aim of experiment: Verification of QC ''' | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (PR1)) | ||
| + | |||
| + | FR11326565 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of P75 (RFP-Generator), F101 (middle linker+ A3C5), P125 (dig. GSY4) into ''E. coli'' XL1-blue and P75 in NEB Turbo ''E. coli'' === | ||
| + | |||
| + | '''Investigator: Niklas, Vivien''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the competent ''E. coli'' cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 1 µl of DNA (6 µl for Ligation F101) was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cells were plated out on Cam-plates/Amp-plate for F101 (inclusive rescue plate) and incubated over the weekend at 30 °C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === QuickChange-PCR of P80 (O23 and O24) and P100 (O19 and O20) === | ||
| + | |||
| + | '''Investigators: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Changing of amino acid | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added) | ||
| + | * Annealing at 55°C | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.25µl | ||
| + | |O23/O24 and O19/O20 | ||
| + | |- | ||
| + | |0,5 µl | ||
| + | |dNTP-mix | ||
| + | |- | ||
| + | |1 µl | ||
| + | |1:100 P80 / P100 | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Pfu Ultra II-buffer | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |Pfu Ultra II Polymerase | ||
| + | |- | ||
| + | |19.25 µl | ||
| + | |dd H<sub>2</sub>O | ||
| + | |- | ||
| + | |4 x 25 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preperative gelelektrophoresis of F102 (PCR of BirA) + phosphorylation and ligation into pSB1C3 === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of experiment:''' Blunt-Ligation of BirA into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *The whole PCR-product was applied to a 1% agorose gel. After runnig the gel two lines where visible: one at 1000Bp and one at ~500Bp. | ||
| + | |||
| + | *The 1000 Bp band was cut out and purified with the Gelextraction kit according to manufacturer's protocoll. Concentration: 41,5ng/µl. | ||
| + | |||
| + | *The DNA was phosphorylated as follows: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |DNA (~1pmol) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Ligase buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4 Polynukleotide-Kinase (for phosphorylation) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *The batch was incubated for 20 min at 37°C and then the enzyme was inactivated for 10 min at 75°C. | ||
| + | |||
| + | *The phosphorylated DNA was then ligated into prepared (HincII-digested) pSB1C3 as follows: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2,3 µl | ||
| + | |Vector | ||
| + | |- | ||
| + | |7,7 µl | ||
| + | |Insert (BirA) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Ligase Buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4 Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *next steps: Transformation into E.coli Xl-1-blue. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Digestion of P78, P123 (for BAP-Nanoluc-construct),P122 (IgKappa for CMV-IgKappa-construct), P89, P114 (for CMV-EGFR-Nanoluc-StrepTag-PolyA-construct) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of experiment:''' Getting of BAP-Nanoluc-construct, CMV-IgKappa-construct and CMV-EGFR-Nanoluc-StrepTag-PolyA-construct. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * DNA was digested according to the following table: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3 µg (for P78, P123 and P122) and 1,5 µg (for P89, P114) | ||
| + | |DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1,5 µl of NgoMIV and PstI | ||
| + | |for P78 | ||
| + | |- | ||
| + | |1,5 µl of AgeI and PstI | ||
| + | |for P123 | ||
| + | |- | ||
| + | |1,5 µl of XbaI and PstI | ||
| + | |for P122 | ||
| + | |- | ||
| + | |1,5 µl of AgeI and PstI | ||
| + | |for P189 | ||
| + | |- | ||
| + | |1,5 µl of NgoMIV and PstI | ||
| + | |for P123 | ||
| + | |- | ||
| + | |up to 50µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' Concentrations: P78 now F106: 2,7 ng/µl, P89 now F107: 14,4 ng/µl. Gelextractions of P114, P122 and P123 didn't contain DNA. (To few DNA available) | ||
| + | |||
| + | *next steps: Retransformation of verified clones (P114, P89) into Xl-1-blue for amplification of plasmid. | ||
| + | Repetition of digestion of P122 and P123 with more DNA (10µg!), then Gel with 100Bp ladder! | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Repetition of digestion of P123 (BAP for BAP-Nanoluc-construct) and P122 (IgKappa for CMV-IgKappa-construct) === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of experiment:''' Getting of BAP-Nanoluc-construct, CMV-IgKappa-construct | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * DNA was digested according to the following table: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µg → 20µl | ||
| + | |DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1,5 µl of AgeI and PstI | ||
| + | |for P123 | ||
| + | |- | ||
| + | |1,5 µl of XbaI and PstI | ||
| + | |for P122 | ||
| + | |- | ||
| + | |up to 50µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Incubation at 37°C over night. | ||
| + | |||
| + | *next steps: Gel with 100Bp ladder and Gelex. Then Ligation with prepared vectors from former digestion (F106 an F63) | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 9, July 11–17= | ||
| + | <div class="week" id="Week_9"> | ||
| + | =='''Monday, July 11'''== | ||
| + | <div class="general"> | ||
| + | |||
| + | === MiniPreps of pSb1C3-Middle Linker+A3C5, -BM40, -CMV-EGFR Retrafo, Streptactin in QC pASK75 === | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim of the experiment:''' Plasmid preparation, analytic digestion, sequencing | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | |||
| + | * Analytic digestion with 0.25 ''µ''L EcoRI, 0.25 ''µ''L PstI (HF) for pSB1C3 plasmids and 0.5 µl NdeI for pASK75-Streptactin, 1 ''µ''L SmartCut Buffer, 3 ''µ''L plasmid DNA, 5.5 ''µ''L H<sub>2</sub>O | ||
| + | |||
| + | * 5 ''µ''L on 1% agarose gel electrophoresis | ||
| + | |||
| + | * plasmid assignment was tricky because of confusion during the weekend Minipreps | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * pASK75-Streptactin clone 1 and 2 were not visible | ||
| + | |||
| + | * verified Plasmid names are available after sequencing | ||
| + | |||
| + | * Lane 1: 5 µl 1 kb Ladder | ||
| + | |||
| + | * Lane 2: 5 ''µ''L Plasmid F101 K1 (seems to be pSB1C3-RFP-Generator) | ||
| + | |||
| + | * Lane 3: 5 ''µ''L Plasmid F101 K2 (seems not to be pSB1C3-RFP-Generator) | ||
| + | |||
| + | * Lane 4: 5 µl 100 bp Ladder | ||
| + | |||
| + | * Lane 5: 5 ''µ''L P125 K1 (seems not to be pSB1C3-RFP-Generator) | ||
| + | |||
| + | * Lane 6: 5 ''µ''L P125 K2 (seems to be pSB1C3-RFP-Generator) | ||
| + | |||
| + | |||
| + | [[File:Muc16_160711_analytic_digestion_cg.jpeg|500px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Preperative digestion of NanoLuc + StrepTag + Poly-A (F112) and cloning behind CMV-EGFR (F107) === | ||
| + | |||
| + | '''Investigator: Christoph, Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' preperative digestion for cloning of the construct behind of CMV-EGFR | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 15 µl P115 (NanoLuc + StrepTag + Poly-A), 1 µl NgoMiV, 1 µl PstI, 3 µl CutSmart buffer, 10 µl H2O | ||
| + | |||
| + | * Incubation for 2 h at 37°C | ||
| + | |||
| + | * 1% gel electrophoresis | ||
| + | |||
| + | Photo | ||
| + | |||
| + | * gel extraction according to SOP | ||
| + | |||
| + | * Ligation of 2 µl F107 (AgeI, PstI) + 8 µl F112 (NgomIV, PstI) with 2 µl Ligase buffer, 7 µl ddH<sub>2</sub>O, 1 µl T4-Ligase for 40 minutes | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate). | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of Nanoluc behind BAP === | ||
| + | |||
| + | '''Investigator: Christoph, Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Cloning of Nanoluc behind BAP based (testing fast cloning strategy) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 5 µl P78 (Nanoluc), 0.5 µl NgoMiV, 0.5 µl PstI, 1 µl CutSmart buffer, 2.5 µl H2O, 0.5 SacI = F108 | ||
| + | * 4 µl P123 (pSB1C3-BAP), 0.5 µl AgeI, 0.5 µl PstI, 1 µl CutSmart buffer, 3 µl H2O, 1 µl FastAP = F109 | ||
| + | |||
| + | * Incubation for 2 h at 37°C, 15 min heat inactivation | ||
| + | |||
| + | * Ligation: 2 µl F108 (Nanoluc) + 8 µl F109 (BAP), 2 µl buffer, 1 µl Ligase, 7 µl H2O | ||
| + | |||
| + | *Transformation: | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate). | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Gel extraction of IgKappa (F111) and cloning behind CMV in pSB1C3 (F63) === | ||
| + | |||
| + | '''Investigator: Christoph, Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' gel extraction after digestion for cloning of the construct behind of CMV | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 1% gel electrophoresis | ||
| + | |||
| + | Photo | ||
| + | |||
| + | * gel extraction according to SOP | ||
| + | |||
| + | * Ligation of 6.1 µl F111 ( XbaI, PstI) + 3.9 µl F63 (SpeI, PstI) with 2 µl Ligase buffer, 7 µl ddH<sub>2</sub>O, 1 µl T4-Ligase for 40 minutes | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate). | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Retrafo of P123 (digest. GSY3-BAP) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 1 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate). | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Digestion of quickchanged mRuby (without StrepTag) and transformation of ''E. coli'' XL1-Blue afterwards === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the PCR-product is digested by 1 µl DpnI (cuts methylated DNA) | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate). | ||
| + | |||
| + | * next steps: 4 Minipreps, test digestion, Sequencing | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === QuickChange-PCR of P100 (O23 and O23) === | ||
| + | |||
| + | '''Investigators: Julian, Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Changing of amino acid in mRuby3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added) | ||
| + | * Annealing at 55°C | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.25µl | ||
| + | |O23/O24 | ||
| + | |- | ||
| + | |0,5 µl | ||
| + | |dNTP-mix | ||
| + | |- | ||
| + | |1 µl | ||
| + | |1:100 P100 | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Pfu Ultra II-buffer | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |Pfu Ultra II Polymerase | ||
| + | |- | ||
| + | |19.25 µl | ||
| + | |dd H<sub>2</sub>O | ||
| + | |- | ||
| + | |4 x 25 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Repeated digestion of P75 (RFP-Generator) with HincII + dephosphorylation === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment:''' Blunt-Ligation of BirA into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Digestion of P75 (RFP-Generator) with HincII + dephosphorylation | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA (P75) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |HincII | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Fast AP (for dephosphorylation) | ||
| + | |- | ||
| + | |11 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Digestion was incubated for 2 hours at 37°C | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Ligation of phosphorylated F102 (PCR of BirA) into F113 (RFP-Generator P75 digest. HincII) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment:''' Blunt-Ligation of BirA into pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *The phosphorylated DNA (prepared on July 8th) was ligated into F113 (HincII-digested RFP-Generator) as follows: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |4,5 µl | ||
| + | |Vector F113 | ||
| + | |- | ||
| + | |5,7 µl | ||
| + | |Insert (BirA) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Ligase Buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4 Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *next steps: Transformation into E.coli NEB Turbo. | ||
| + | </div> | ||
| + | |||
| + | ==Tuesday, July 12== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | ===Transformation of BirA–RFP generator ligation (F114) and Re-Transformation of P114=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Transform NEB Turbo cells | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Followed standard transformation protocol (SOP) using 1 μL and 10 μL of P114 and F114, respectively | ||
| + | |||
| + | * Incubated at 37 °C for 1:10 h | ||
| + | |||
| + | * Plated on LB Cam (50 μL + rescue) | ||
| + | |||
| + | * Incubated at 37 °C (start 12:30) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | ===Arabinose Induction Experiment: pBAD–GFP=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Test different induction concentrations of arabinose | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Of 30 °C o/n culture, filled 5 mL each into four 10 mL culture tubes | ||
| + | |||
| + | * Induced with 0, 0.1, 1, and 10 mM final conc. of arabinose | ||
| + | |||
| + | * Took 0.5 mL sample each and stored at 4 °C | ||
| + | |||
| + | * Incubated 5 mL cultures at 30 °C for 4 h | ||
| + | |||
| + | * Took another 0.5 mL sample each and stored at 4 °C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Picking Colonies of F101 (P69 Middle Linker – P112 A3C5)=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Pick transformed colonies | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Picked 3 colonies off plate with transformed cells | ||
| + | |||
| + | * Let grow at 37 °C in LB Amp (start 11:50 am) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Picking Colonies of F123 (BAP)=== | ||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Pick transformed colonies | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Picked 2 colonies off plate with transformed cells | ||
| + | |||
| + | * Let grow at 37 °C in LB Cam (start 17:00) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | ===Verification of succesful Quickchange of P141 (pASK75 with Streptactin)=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the Plasmid (5 µl) is digested by 0.5 µl NdeI (Addiition of 1 µl cut smart and 3.5 µl ddH2O) | ||
| + | |||
| + | * since the quickchange should eliminate the second Nde-restriction site, there should be one band on the gel | ||
| + | |||
| + | '''result:''' | ||
| + | |||
| + | * there is no band on the gel (the experiment was repeated multiple times showing the same results) | ||
| + | |||
| + | * '''therefore new primers are designed and ordered''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Refolding of Streptavidin=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the sample (pelett dissolved in 6M GdmCl)was spun down (4°C, 20 mins, 18,000 rpm) | ||
| + | |||
| + | * the supernatant was transferred carefully into a falcon tube and the pellet was cast away | ||
| + | |||
| + | * the supernatant was added dropwise to 1 X PBS (500 ml) and stirred over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Colony PCR of F110 (BAP+Nanoluc)=== | ||
| + | |||
| + | '''Investigator: Vivien, Niklas''' | ||
| + | |||
| + | '''Aim:''' Amplification | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * samples were prepared with 5 µL Q5-Mastermix, 1 µl VF2-, 1 µL VR-Primer, 2 µL ddH20 | ||
| + | |||
| + | * four clones were picked and added to the samples respectively | ||
| + | |||
| + | * PCR- cycles according to SOP | ||
| + | |||
| + | * Lane 1: 1 kb Ladder, Lane 2-5: clones 1-4, 10 µl each | ||
| + | |||
| + | [[File:Muc16_ColonyPCR_F110_1307.jpeg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion and gelelectrophoresis of P37 and P26 === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment: Preparative digestion and gelelectrophoresis of P137 (BM40)and P26 (CMV)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | *P137: EcoRI and XbaI | ||
| + | |||
| + | *P26: EcoRI and SpeI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |23/7 µl | ||
| + | |Plasmid DNA P137/P26 | ||
| + | |- | ||
| + | |19/35 µl | ||
| + | |ddH2O P137/P26 | ||
| + | |- | ||
| + | |5 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |each enzyme(10 U/µl) | ||
| + | |- | ||
| + | |50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | *Preparative gelelectrophoresis was performed at 90 V for 60 min | ||
| + | |||
| + | *The whole PCR-product was applied to a 1% agorose gel. After runnig the gel two lines where visible: one at 1000Bp and one at ~500Bp. | ||
| + | |||
| + | *The 600/2100 bp band (P26/P137) was cut out and purified with the Gelextraction kit according to manufacturer's protocoll. Concentration: 20.4/33.1 ng/µl. (F115/F116) | ||
| + | |||
| + | [[File:Muc16_P137-P26_1207.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligation of F115 into F116 === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment:''' Ligation of F115 (CMV) into F116 (BM40 in pSB1C3) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |4,3 µl | ||
| + | |Vector F116 (BM40 in pSB1C3) | ||
| + | |- | ||
| + | |5,7 µl | ||
| + | |Insert F115(CMV) | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Ligase Buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4 Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *next steps: Transformation into E.coli NEB Turbo. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of F117 (CMV in pSB1C3 with BM40), F118 (QC P100), F119 (QC P80) and P26 (CMV; Retrafo) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of CMV in pSB1C3 with BM40 (F117), QC P100 (F118), QC P80 (F119) and P26 in NEB Turbos(F117)/XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. coli NEB Turbo (F117)/XL1 blue (rest) according to protocol + rescue | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | </div> | ||
| + | |||
| + | |||
| + | ==Wednesday, July 13== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === QuickChange-PCR of P80 (O23 and O24) === | ||
| + | |||
| + | '''Investigators: Christoph''' | ||
| + | |||
| + | '''Aim of the experiment:''' Changing of amino acid in mRuby3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added) | ||
| + | * Annealing at 55°C | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.25µl | ||
| + | |O23/O24 | ||
| + | |- | ||
| + | |0,5 µl | ||
| + | |dNTP-mix | ||
| + | |- | ||
| + | |1 µl | ||
| + | |1:100 P80 | ||
| + | |- | ||
| + | |2.5 µl | ||
| + | |Pfu Ultra II-buffer | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |Pfu Ultra II Polymerase | ||
| + | |- | ||
| + | |19.25 µl | ||
| + | |dd H<sub>2</sub>O | ||
| + | |- | ||
| + | |4 x 25 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | Cycling was performed according to SOP. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === MiniPrep of Middle Linker-A3C5 construct === | ||
| + | |||
| + | '''Investigator: Christoph ''' | ||
| + | |||
| + | '''Aim of the experiment:''' MiniPrep two colonies of Middle Linker-A3C5 construct | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | * Both plasmids were sent to sequencing without analytical digestion because of the short lenght | ||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Cloning of F120 (CMV-IgKappa) and F121 (CMV+EGFR / Nanoluc+StrepTag+PolyA) === | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligations of F120 (CMV-IgKappa) and F121 (CMV+EGFR / Nanoluc+StrepTag+PolyA) and transformation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ligation mixtures were prepared as following: | ||
| + | * F120: 5 μL F63, 5 µL F111, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * F121: 3 μL F107, 7 μL F112, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase | ||
| + | * Ligation was incubated for 30 min. at room temperature | ||
| + | * transformation was performed according to SOP (with 100 µl cells and 20 µl ligation mix) and plating in LB+Cam agar | ||
| + | |||
| + | </div> | ||
| + | |||
| + | ==Thursday, July 14== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===MiniPreps of Overnight Cultures=== | ||
| + | |||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Prep plasmid DNA of P26, F110, F117, F114, P114, and P118 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep performed according to manufacturer's instructions with Qiagen kit | ||
| + | |||
| + | * Prior to eluting, 50 μL EB buffer was applied to the membrane, and the columns were heated at 37 °C for 7 min | ||
| + | |||
| + | '''Results:''' Plasmid DNA solutions of the following concentrations: P26 (a: 55.1 ng/μL; b: 48.1 ng/μL); F110 (K2: 150.1 ng/μL; K3: 173.0 ng/μL); F117 (a: 257.7 ng/μL; b: 159.0 ng/μL; CMV/BM40: 133.2 ng/μL); F114 (83.1 ng/μL); P114 (a: 26.1 ng/μL; 170.7 ng/μL); P118 ('''F118?''') (a: 248.0 ng/μL; 53.2 ng/μL; 241.3 ng/μL). F110 (K2: FR12904474; K3: FR12904473) and F117 (a: FR12904477; b: FR12904476; CMV/BM40: FR12904475) were sent for sequencing with VF2. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === DpnI Digest of mRuby3 QC and Transformation=== | ||
| + | |||
| + | '''Investigator: Vivien''' | ||
| + | |||
| + | '''Aim:''' Digest original-sequence plasmid and transform obtained pure quick-changed plasmid | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * QC was digested with DpnI (40 μL QC, 5 μL 10× CutSmart buffer, 3.5 μL ddH<sub>2</sub>O, 1.5 μL DpnI = 50 μL) at 37 °C for 2 h | ||
| + | |||
| + | * Digest (10 μL) was directly transformed into Turbo cells following SOP (including 1 h incubation time prior to plating due to chloramphenicol resistance) | ||
| + | |||
| + | * Plated 50 μL on LB Cam + rescue plate | ||
| + | |||
| + | * Incubated at 37 °C | ||
| + | |||
| + | '''Result:''' two plates; remaining digest stored at 4 °C in styrofoam tube rack | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, July 15'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Inoculation of mRuby Quick change, F120 (CMV+IgKappa) and F121 (CMV+EGFR/Nanoluc+StrepTag+PolyA) preculture and miniprep === | ||
| + | |||
| + | '''Investigator: Niklas, Clemens''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 6 colonies were picked from plate | ||
| + | * Cells were inoculated in 5 mL LB medium (Cam 1:1000) | ||
| + | * Fragments were minipreped following manufacturer's protocol (QiaPrep, Qiagen) | ||
| + | * Fragments were renamed receiving a P number | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Quickchange of P88 (pASK75(Streptactin)) with O-029 and O-030=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim:''' Quickchange pASK75(Streptactin) to erase one NdeI restriction site | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * For quickchange two seperate batches were prepared: | ||
| + | * 1. batch: 0.7 µL O-029, 0.5 µL dNTP mix, 2.5 µL buffer, 0.5 µL Pfu ultra polymerase, 1.5 µL P88 | ||
| + | * 2. batch: 0.7 µL O-030, 0.5 µL dNTP mix, 2.5 µL buffer, 0.5 µL Pfu ultra polymerase, 1.5 µL P88 | ||
| + | * first stage of quickchange PCR was performed as following: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial Denaturation||98||30 | ||
| + | |- | ||
| + | |rowspan = "3"|15 cycles||98||10 | ||
| + | |- | ||
| + | |55||30 | ||
| + | |- | ||
| + | |68||5 | ||
| + | |- | ||
| + | |final Extension||68||5 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | * PCR batches were mixed and 0.5 µL pfu ultra polymerase was added | ||
| + | * Second stage of quickchange PCR was performed as in the following: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial Denaturation||98||30 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |55||30 | ||
| + | |- | ||
| + | |68||5 | ||
| + | |- | ||
| + | |final Extension||68||5 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of P79 (Nanoluc) into P147 (pSB1C3(BAP))=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 7 µL P79 were digested with NgOMIV and PstI | ||
| + | * 10 µL P147 were digested with AgeI and PstI | ||
| + | * Digestion products were run on a 1% agarose gel | ||
| + | * Both fragments could be cut from the gel | ||
| + | * Fragments were purified by gelectraction following manufacturer's protocol (Qiagen) | ||
| + | |||
| + | *Fragments were renamed: | ||
| + | ** digest P78= F123 | ||
| + | ** digest P147= F124 | ||
| + | * 9,5 µL F124 and 0.5 µL F123 were ligated following manufacturers protocol (T4 DNA ligase, Thermo scientific) | ||
| + | * NEB Turbo were transformed with 7 µL of F125 (ligation F123+F124) following SOP | ||
| + | * Cells were plated on CAM LB medium and placed in the incubator at 37°C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of P145 (Middle Linker +A3C5) and P146 (Middle Linker +A3C5) into F70 (Vector fragment pSB1C3)=== | ||
| + | |||
| + | '''Investigator: Niklas, Clemens''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Ligation: | ||
| + | ** 2,5 µL P145 and 1 µL P146 were pooled and digested with NgOMIV and SpeI for 1h at 37°C. | ||
| + | ** Full digestion was ligated into 1 µL F70 | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Add 1 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 60 µl of the cell suspension was plated on one chloramphenicol plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a chlorampenicol plate (=rescue plate). | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Test digestion of P164 (BirA blunt end ligation) (after miniprep) and P161, P162, and P163 (all QC mRuby)=== | ||
| + | |||
| + | '''Investigator: Niklas, Clemens''' | ||
| + | |||
| + | '''Aim:''' Test digestion to see which plasmids contain the right insert | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 150 ng of plasmid DNA were digested with EcoRI and PstI following SOP | ||
| + | * The whole batch was applied to a 1% gel to check for cut inserts. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | FOTO | ||
| + | |||
| + | * Blunt end ligation seems to not have worked (P164) | ||
| + | * P161, P162 and P163 all still carry the insert | ||
| + | |||
| + | Sequenced: P161= FR12904472 (NA) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Harvesting of Streptavidin=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * After precipitation in 70 % ammonium sulfate the soluton was spun down in the centrifuge (4°C, 11000 rpm, 40 mins) | ||
| + | * The supernatant was discarded, and the pellet was dissolved in 2.2 M ammonium sulfate (18 mL) | ||
| + | * the sample was spun down in the centrifuge (4°C, 15000 rpm, 30 mins) | ||
| + | * the pellet was dissolved in 1X PBS(16 mL) | ||
| + | * next steps: last centrifugation and dialysis against Tris/Hcl (see plan on board) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 10, July 18–24= | ||
| + | <div class="week" id="Week_10"> | ||
| + | |||
| + | =='''Monday, July 18'''== | ||
| + | <div class="receptor"> | ||
| + | |||
| + | === Inoculation of precultures for miniprep of F125 (pSB1C3(BAP-NanoLuc)) and F122 (pSB1C3(Middlelinker-A3C5))=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | * Miniprep, analytical digestion, analytical gel, sequencing | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Three colonies were picked from each plate (F125, F122) | ||
| + | * Inoculation was performed according to SOP | ||
| + | * Cells grew for 5h | ||
| + | ** Cells onlygrew in cultures with F125 (In cultures with F122 there was no pelet) | ||
| + | * Miniprep was performed according to manufactrer's protocol | ||
| + | * Samples were renamed: | ||
| + | ** F125 K1: P168 | ||
| + | ** F125 K2: P169 | ||
| + | ** F125 K3: P170 | ||
| + | |||
| + | * Analytical digestion: | ||
| + | ** Each plasmid was digested with EcoRI and PstI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P168/P169/P170 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI(10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI (20 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Foto: | ||
| + | |||
| + | *Lane 1: 1kb ladder | ||
| + | *Lane 2: P168 | ||
| + | *Lane 3: P169 | ||
| + | *Lane 4: P170 --> right signals (2000 bp, ~600 bp(BAP-NanoLuc) | ||
| + | |||
| + | |||
| + | *Send for sequencing: | ||
| + | **P170: FR12904468 (VF2) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion of P149 , P150 (both pSB1C3(CMV-IgKappa)), P151, P152 (both pSB1C3(CMV-EGFR-NanoLuc-StrepTag-polyA)), P153, P154 (both QC pSB1C3(mRuby3))=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | Analytical digeston to find out if the cloning was successful | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * measure concentrations of Plasmids (see inventary list) | ||
| + | |||
| + | * Analytical digestion: | ||
| + | ** Each plasmid was digested with EcoRI and PstI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Plasmid DNA P149/ P150/ P151/ P152/ P153/ P154 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI(10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI (20 U/µl) | ||
| + | |- | ||
| + | |5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Foto: | ||
| + | |||
| + | *Lane 1: 1kb ladder | ||
| + | *Lane 2: P149 --> right signals (2000bp; 730bp) | ||
| + | *Lane 3: P150 --> right signals (2000bp; 730bp) | ||
| + | *Lane 4: P151 | ||
| + | *Lane 5: P152 --> right signals (2000bp; 1626bp) | ||
| + | *Lane 6: P153 --> very weak signal | ||
| + | *Lane 7: P154 --> right signals (2000bp; 700bp) | ||
| + | |||
| + | *Send for sequencing: | ||
| + | ** P150: FR12904471 (VF2) | ||
| + | ** P152: FR12904470 (VF2) | ||
| + | **P154: FR12904469 (VF2) | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of QC of P88 (pASK75(Streptactin)) === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | * See if quickchange has worked | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * NEB Turbo cells were transformed with 10 µl of QC P88 according to SOP | ||
| + | * Cells were plated on LB medium (Amp) and incubated at 37°C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Digestion of P165 (pSB1C3(CMV-BM40) and cloning of F112 (Nanoluc-StrepTag-polyA) into P165 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | * Create biobrick: CMV-BM40-Nanoluc-StrepTag-polyA | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Preparative digestion batch was prepared as in the following: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA P156 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI(10 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI(20 U/µl) | ||
| + | |- | ||
| + | |10 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestion was seperated by gelelectrophoresis | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Foto: | ||
| + | |||
| + | * Lane 1: 1kb ladder | ||
| + | * Lane 2: F126 (digest P165) --> right length | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Fragment was cut from the gel and purified by gelextraction which was performed by manufacturer's protocol (Qiagen) | ||
| + | |||
| + | * F126 was ligated with F112 | ||
| + | |||
| + | Ligation batch: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Vector DNA F126 | ||
| + | |- | ||
| + | |18 µl | ||
| + | |Insert DNA F112 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |T4 ligation buffer (10x) | ||
| + | |- | ||
| + | |6 µl | ||
| + | |T4 ligase | ||
| + | |- | ||
| + | | µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Ligation was incubated at 37°C for 30 min | ||
| + | * NEB Tubros were transformed with F127 (lig. F112+F126) | ||
| + | * Cells were plated on LB medium (Cam) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, July 19'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | |||
| + | ===Inoculation of Precultures of F127 (pSB1C3(CMV-BM40-Nanoluc-StrepTag-polyA))=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim:''' Prepare cells for miniprep of plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * six colonies were picked and inoculated in LB medium (CAM) | ||
| + | * Plates were incubated on a shaker at 37°C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * cellgrowth ? | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Reamplifiying Streptactin from GSY6=== | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim:''' Reamplifiying Streptactin from GSY6 for further processing in QC reaction (delete NdeI site from pASK) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * PCR reaction was prepared according to SOP | ||
| + | * Cycling steps were taken from the QC protocol because no further cycler was free | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * will be available after DpnI digestion and transformation | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === QuickChange-PCR of pASK75 with O29 and O30 === | ||
| + | |||
| + | '''Investigator: Christoph''' | ||
| + | |||
| + | '''Aim:''' Deleting NdeI from pASK (P10) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * QC-PCR was performed according to SOP | ||
| + | * Primers: O-029 and O-030 | ||
| + | * Annealing at 55°C | ||
| + | * Elongation time: 5 min | ||
| + | * 2-step protocol was applied | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Miniprep of E. coli Xl1-Blue P171, P172 and P173 (pSB1C3(CMV-BM40-Nanoluc-StrepTag-polyA)) === | ||
| + | |||
| + | '''Investigator: Javier''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extracting P171, P172 and P173 (CMV-BM40-Nanoluc-StrepTag-polyA) from E.coli XL-1-blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN) | ||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P171 | ||
| + | </td><td>75,4 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P172 | ||
| + | </td><td>108,2 | ||
| + | <tr> | ||
| + | <td>P173 | ||
| + | </td><td>66,4 | ||
| + | </td></tr></table> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Transformation of F136 (A3C5+Middle-Linker), F137 (EGFR+mRuby-Strep), F138 (mRuby3-Strep+PolyA) and P161 (QC mRuby-Strep) === | ||
| + | |||
| + | '''Investigator: Javier''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of A3C5+Middle-Linker (F136), EGFR+mRuby-Strep (F137), mRuby3-Strep+PolyA (F138) and QC mRuby-Strep (P161) XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent E. coli XL1 blue according to protocol + rescue | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB Cam) in incubator for further processing (37 °C) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Colony PCR of F127 (CMV-BM40-Nanoluc-StrepTag-polyA)=== | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim:''' See if cloning was successfull | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * samples wwere prepared with 5 µL Q5-Mastermix, 1 µl VF2-, 1 µL VR-Primer, 2 µL ddH20 | ||
| + | |||
| + | * six clones were picked and added to the samples respectively | ||
| + | |||
| + | * PCR- cycles according to SOP (Annealing-temp: 66°C; Ampflification time: 1.5min) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Lane 1: 1 kb Ladder | ||
| + | * Lane 2: F127 K1 | ||
| + | * Lane 3: F127 K2 | ||
| + | * Lane 4: F127 K3 | ||
| + | * Lane 5: F127 K4 | ||
| + | * Lane 6: F127 K5 | ||
| + | * Lane 7: F127 K6 | ||
| + | |||
| + | [[File:Muc16_ColonyPCR_F117_1907.jpeg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | |||
| + | ===Preparative digestion of P161 (pSB1C3(mRuby3-StrepTag)), P73 (pSB1C3((PolyA)), P96 ((pSB1C3(EGFR-TMD)), P69 (pSB1C3(Middle-linker)) and P112 (pSB1C3(A3C5)) === | ||
| + | |||
| + | '''Investigator: Clemens, Christoph, Javier''' | ||
| + | |||
| + | '''Aim:''' Create biobricks | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | * Preparative digestion of P161 and P73 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl/ 20 µl | ||
| + | |Plasmid DNA P161/ P73 | ||
| + | |- | ||
| + | |3 µl/ 3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |EcoRI/ EcoRI | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |SpeI/ XbaI | ||
| + | |- | ||
| + | |10 µl/ 5 µl | ||
| + | |ddH2O/ ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Preparative digestion of P96 and P161 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl/ 15 µl | ||
| + | |Plasmid DNA P96/ P161 | ||
| + | |- | ||
| + | |3 µl/ 3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |PstI/ PstI | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |AgeI/ NgOMIV | ||
| + | |- | ||
| + | |15 µl/ 10 µl | ||
| + | |ddH2O/ ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Preparative digestion of P69 and P112 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |20 µl/ 15 µl | ||
| + | |Plasmid DNA P69/ P112 | ||
| + | |- | ||
| + | |3 µl/ 3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |EcoRI/ EcoRI | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |AgeI/ NgOMIV | ||
| + | |- | ||
| + | |5 µl/ 10 µl | ||
| + | |ddH2O/ ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated at 37°C for 2h | ||
| + | * Samples were applied to a 1% agarose gel (40 min; 90V) | ||
| + | * Samples were cut from the gel afterwards: | ||
| + | * Renamed: | ||
| + | ** digest P69: F128 | ||
| + | ** digest P112: F129 | ||
| + | ** digest P161: F130 | ||
| + | ** digest P96: F131 | ||
| + | ** digest P161: F132 | ||
| + | ** digest P73: F133 | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Fotos | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Gelextraction was performed after manufacturers protocol (Qiagen) | ||
| + | * After gelextraction concentrations were measured: | ||
| + | |||
| + | |||
| + | * Ligation: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |xx µl | ||
| + | |Vektor F129 | ||
| + | |- | ||
| + | |xx µl | ||
| + | |Insert F128 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |x µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |xx µl | ||
| + | |Vektor F131 | ||
| + | |- | ||
| + | |7xx µl | ||
| + | |Insert F130 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |x µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |xx µl | ||
| + | |Vektor F133 | ||
| + | |- | ||
| + | |7xx µl | ||
| + | |Insert F132 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |x µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Ligation was performed according to SOP | ||
| + | * Xl1 blue cells were transformed with the Ligations according to SOP | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, July 20'''== | ||
| + | <div class="general"> | ||
| + | |||
| + | ===Analytical digestion and gelelectrophoresis of P171-P173 & QC of P10 === | ||
| + | |||
| + | '''Investigator: JB, MT''' | ||
| + | |||
| + | '''Aim of experiment:''' Check if everything is fine. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digest P171-P173 with XbaI + SpeI and QC of P10 with NedI and incubate 1 h at 37 °C | ||
| + | * Run 1 % agarose-gel at least 40 min at 100 V | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_digestion_P171_P172_P173_QCP10.jpeg|500px]] | ||
| + | |||
| + | Lane1: 1Kb ladder; Lane2: P171; Lane3: P172; Lane4: P173; Lane5: QC of P10 | ||
| + | |||
| + | => P171 was sent for sequencing: FR12904466 | ||
| + | => NdeI test digestion shows two signals although there should have been just one | ||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Preparation of GSY 6; F134; F135 for ligation=== | ||
| + | |||
| + | '''Investigator: JB, MT ''' | ||
| + | |||
| + | '''Aim of experiment:''' Prepare GSY6 for Ligation. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Digestion of GSY6 with SpeI + HindIII | ||
| + | * Preparative 1 % Agarose gel 100 V for at least 40 min. | ||
| + | * Gel extraction of GSY6, F134, F135 by Quiagen kit. | ||
| + | |||
| + | '''Results:''' | ||
| + | * GSY6 -> F139 : 2.5 ng/µL | ||
| + | * F134 : 4 ng/µL | ||
| + | * F135 : 10.2 ng/µL | ||
| + | |||
| + | Preparative agarose gel GSY6: | ||
| + | |||
| + | [[File:Muc16_digestion_prep_gel_GSY6.jpeg|300px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | ===Ligation & transformation of F134 + F63; F135 + F139 === | ||
| + | |||
| + | '''Investigator: JB, MT''' | ||
| + | |||
| + | '''Aim of experiment:''' Ligation & transformation of F134 + F63; F135 + F139 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Ligation following SOP(13.02.2013) | ||
| + | * F134 + F63 -> F140 | ||
| + | * F135 + F139 -> F141 | ||
| + | |||
| + | transformation following SOP(13.02.2013) | ||
| + | * F140 on CAM | ||
| + | * F141 on AMP | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | We will see tomorrow, please update this section. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, July 21'''== | ||
| + | <div class="receptor"> | ||
| + | |||
| + | === Miniprep of F136 (K1 & K2) (pSB1C3(Middle linker-A3C5)), F137 (K1 & K2) (pSB1C3(EGFR TMD-mRuby3-StepTag)), F138 (K1 & K2) (pSB1C3(mRuby3-StrepTag-polyA) and retrafo of P161 (pSB1C3(mRuby3-StrepTag)) === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of experiment:''' Get plasmids for analytical digestion and sequencing | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Minipreps were performed according to manufacturers protocol (Qiagen, QiaPrep) | ||
| + | * Samples were renamed afterwards: | ||
| + | ** F136 K1: P174 | ||
| + | ** F136 K2: P175 | ||
| + | ** F137 K1: P176 | ||
| + | ** F137 K2: P177 | ||
| + | ** F138 K1: P178 | ||
| + | ** F138 K2: P179 | ||
| + | ** retrafo of P161: P180 | ||
| + | |||
| + | * Analytical digestion: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Ligation was renamed: F143 | ||
| + | * Digestion products were seperated via gelelectrophoresis | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Foto: | ||
| + | |||
| + | * All plasmids seemed to carry the appropriate insert | ||
| + | |||
| + | Sequencing: | ||
| + | |||
| + | ** P175: FR12904465 (VF2) | ||
| + | ** P177: FR12904464 (VF2) | ||
| + | ** P178: FR12904463 (VF2) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion of P116 (pSB1C3(IgKappa)) === | ||
| + | |||
| + | '''Investigator: Clemens, Christoph''' | ||
| + | |||
| + | '''Aim of experiment:''' cloning of CMV in front of IgKappa signal peptide | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * preparative digestion batch | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA P116 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestion was applied to a 1% agarose gel (45 min; 90V) | ||
| + | * Sample was cut from the gel and gelextraction was performed according to manufacturers protocol | ||
| + | |||
| + | * Ligation of Fx and F115 (digest CMV promotor): | ||
| + | |||
| + | * Ligation: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3.9 µl | ||
| + | |Vektor F142 | ||
| + | |- | ||
| + | |6.1 µl | ||
| + | |Insert F115 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |20 µl | ||
| + | |ddH<sub>2</sub>O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Transformation of XL1blue was performed according to SOP | ||
| + | * Cells were plated on LB medium (CAM) and incubated over night at 37°C | ||
| + | |||
| + | </div> | ||
| + | =='''Friday, July 22'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Analytical digestion of pASK_T7 RBS_LCN2_His=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | Analytical digestion to find out whether there is more than one NdeI restriction site. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Analytical digestion: | ||
| + | **The plasmid was digested with NdeI and HindIII | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |NdeI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |HindIII | ||
| + | |- | ||
| + | |5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * Two bands were visible what means that there is only one NdeI restriction site. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Inoculation of Precultures of F143 (pSB1C3(CMV+IgKappa))=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Prepare cells for miniprep of plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * two colonies were picked and inoculated in LB medium (CAM) | ||
| + | * Plates were incubated on a shaker at 37°C over night. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | ===Harvesting of Streptavidin=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Recombinant expression and purification of streptavidin | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the solution was spun down in the centrifuge (4°C, 15000 rpm, 20 mins) to remove last contamination | ||
| + | * The pellet was discarded and the supernantant was dialysed against 20 mM Tris/HCl (ph 8) witt 50 mM NaCl | ||
| + | * last step to do: steril filtration | ||
| + | |||
| + | * the concentration was determined via UV/VIS: the absorption at 280 nm is 0.469 (1:100 dilution), this equals a concentration of 62 mg/mL (about 16 mL) (mass 54 kDa; Absorbtion coefficient 40455 1/M*cm | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Sequencing of P174(Middle Linker + A3C5) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of P178 (mRubyStrepPolyA) / P180 (mRubyStrep) behind EGFR-TMD in pSB1C3 (P96) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Preparative digestion: | ||
| + | * The Inserts were digested with PstI and NgoMIV | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |7.7 / 8.2 µl (about 3µg) | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgoMIV | ||
| + | |- | ||
| + | |8.3 / 7.8 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * both fragments were ligated with F131 (digest P96 [AgeI; PstI]) | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | * Results: Cloning didn't work. No colonies on plate. Repeat. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class:"receptor"> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of Middle Linker before A3C5 in pSB1C3 (Repeat)=== | ||
| + | |||
| + | '''Investigator: Julian, Niklas''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Preparative digestion: | ||
| + | **The Middle Linker (P69) was digested with EcoRI and AgeI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |30 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |EcoRI | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |AgeI | ||
| + | |- | ||
| + | |12 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * the fragment was ligated with F129(digest P112 [EcoRI; NgOMIV]) | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 11, July 25th - July 31st= | ||
| + | <div class="week" id="Week_11"> | ||
| + | |||
| + | =='''Monday, July 25'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Inoculation of Precultures of F143 (pSB1C3(CMV+IgKappa)), F141new (pASK(Streptactin)), F151 (pSB1C3(middleLinker+A3C5), pASK75_T7-RBS-LCN2-His, F148 (pSB1C3(mRubyStrepPolyA+ CMV-EGFR Signal-Pept)=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Prepare cells for miniprep of plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Three colonies of each were picked and inoculated in LB medium (CAM) respectively (AMP) fpr pASK. | ||
| + | * Plates were incubated on a shaker at 37°C for 6 hours. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===MiniPreps and analytical digestion of F143 cultures=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Prep plasmid DNA of F143 (pSB1C3(CMV + IgKappa)) and checking wheather ligation was successful. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep performed according to manufacturer's instructions with Qiagen kit | ||
| + | |||
| + | * Prior to eluting, 30 μL EB buffer was applied to the membrane since the pellet was quite small, and the columns were heated at 37 °C for 7 min | ||
| + | |||
| + | '''Results:''' | ||
| + | * Concentrations: K1 = 230,4ng/µl, K2 = 162,4 ng/µl | ||
| + | |||
| + | '''Analytical Digestion:''' | ||
| + | |||
| + | * An 20µl batch was prepared for analytical digestion for each of the two clones: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Two bands were visible at 750 Bp which is the size of IgKappa and CMV together. Cloning was successful. Sequence still has to be ckecked though. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of P178 (mRubyStrepPolyA) / P180 (mRubyStrep) behind EGFR-TMD in pSB1C3 (P96). Repetition. === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' See title. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *Digestion of P178 and P180: 7µg of DNA were digested. | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |18 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |2,5 µl | ||
| + | |NgoMIV | ||
| + | |- | ||
| + | |2,5 µl | ||
| + | |PstI | ||
| + | |- | ||
| + | |22 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestion at 37°C for 2,5h. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of NanoLuc-Strep-PolyA (F112) behind CMV-IgKappa in pSB1C3 (P181). === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' See title. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *Digestion of P181: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |2,5 µl | ||
| + | |AgeI (20u/µl) | ||
| + | |- | ||
| + | |2,5 µl | ||
| + | |PstI (20u/µl) | ||
| + | |- | ||
| + | |30 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestion at 37°C for 2,5h. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Digestion and gel-extraction of P178; P180=== | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of experiment:''' Preparation of P178 and P180 for ligation. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Digest P178; P180 with NgoMIV + PstI incubate 1 h at 37 °C | ||
| + | * Run 1 % agarose-gel at least 40 min at 100 V | ||
| + | * Gel extraction by Quiagen kit. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * P178 [NgoMIV +PstI digest] -> F152: 17 ng/µL | ||
| + | * P180 [NgoMIV +PstI digest] -> F153: 8.1 ng/µL | ||
| + | |||
| + | [[File:Muc16_digestion_prep_gel_P178_P180.jpeg|300px]] | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Ligation & transformation of F131 + F152; F131 + F153 === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of experiment:''' Ligation & transformation of F131 + F152; F131 + F153 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Ligation following SOP(13.02.2013) | ||
| + | * F131 + F152 -> F154 | ||
| + | * F131 + F153 -> F155 | ||
| + | |||
| + | Transformation following SOP(13.02.2013) | ||
| + | * F154 E. coli. turbo CAM | ||
| + | * F155 E. coli. turbo CAM | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | We will see tomorrow, please update this section. | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, July 27th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Minipreps of EGFR-TMD - mRuby - Strep - PolyA (F131+F152) and EGFR-TMD - mRuby - Strep (F131+F153) === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Results:''' | ||
| + | Procedure according to protocol (spin kit). Resulting plasmids: | ||
| + | |||
| + | EGFR-TMD - mRuby - Strep - PolyA (F131+F152) -> P196, P197 | ||
| + | EGFR-TMD - mRuby - Strep (F131+F153) -> P198, P199 | ||
| + | |||
| + | Concentrations: | ||
| + | P196: 228 ng/µl | ||
| + | P197: 212 ng/µl | ||
| + | P198: 197 ng/µl | ||
| + | P199: 322 ng/µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, July 28th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Inoculation of Precultures of F141new (pASK(Streptactin))=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Prepare cells for miniprep of plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Three colonies of each were picked and inoculated in LB medium (CAM) respectively (AMP) fpr pASK. | ||
| + | * Plates were incubated on a shaker at 37°C for 6 hours. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ==='''PCR of GSY6'''=== | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Amplification of Streptavidin mutant and Traptactin for further cloning into pASK. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *reaction was performed after SOP (PCR Using Q5® High-Fidelity DNA Polymerase, NEB) | ||
| + | * 2ng Plasmid were used for amplification (1 µL) | ||
| + | * Primers: VR and VF2 | ||
| + | |||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial Denaturation||98||30 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |66||30 | ||
| + | |- | ||
| + | |72||90 | ||
| + | |- | ||
| + | |final Extension||72||5 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | * PCR product was purified accoding to manufacturer's protocol (PCR puification kit, Qiagen) | ||
| + | * DNA was eluted 40 µL EB buffer | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Digestion of pASK === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim:''' Preparing pASK for ligation with Streptavidin mutant and Traptactin. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |20 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1,5 µl | ||
| + | |NdeI | ||
| + | |- | ||
| + | |1,5 µl | ||
| + | |HindIII | ||
| + | |- | ||
| + | |22 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Cloning of Traptactin and Streptavidin mutant into pASK (P191). === | ||
| + | |||
| + | '''Investigator: Luisa''' | ||
| + | |||
| + | '''Aim''': Preperation of the plasmids for Streptavidin and Traptactin expression in E.coli. | ||
| + | |||
| + | '''Procedure''': | ||
| + | |||
| + | * Cutting out the crucial bands from the gel containing the PCR-product and the digested plasmid (P190) and purifiing them with the Gelextraction Kit according to the protocoll. '''First Gel failed: Very small band with wrong length...--> Repetition of digestion with P190.''' | ||
| + | |||
| + | * The PCR product of GSY6 was extracted from the gel (Band at ~800 Bp) with the Gelextraction Kit and then digested with NdeI and HindIII to get the fragments Streptavidin m1 and Traptactin. Inactivation was performed at 80°C for 10 min. | ||
| + | |||
| + | * Concentrations: 33,9ng/µl (GSY6), | ||
| + | |||
| + | |||
| + | * Ligation... | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of NanoLuc-Strep-PolyA (F112) behind CMV-IgKappa in pSB1C3 (P182). Repetition. === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim:''' See title. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *Digestion of P182: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1,5 µl | ||
| + | |AgeI (20u/µl) | ||
| + | |- | ||
| + | |1,5 µl | ||
| + | |PstI (20u/µl) | ||
| + | |- | ||
| + | |9 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestion at 37°C for 2.5h. | ||
| + | * Take care: P182 is not sequenced | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Retrafos of P178 (mRuby-Strep-PolyA) and P181 (CMV + IgKappa)=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Results:''' | ||
| + | Transformation according to protocol, plates were incubated at 37°C overnight. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Dialysis of eGFP=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Preparation of eGFP for following biotinylation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * eGFP was poured in a dialysis hose (cut-off 14 kDa) | ||
| + | * The hose was then placed in 4 L ice cold borate buffer 50 mM pH 8.85 (no salt) | ||
| + | * The dialysis took place at 4°C over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Sequencing of P160 (NanoLuc+StrepTag+Poly-A) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, July 29th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Minipreps of EGFR-TMD - mRuby - Strep - PolyA (F131+F152) and EGFR-TMD - mRuby - Strep (F131+F153) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Procedure according to protocol (spin kit). Resulting plasmids: | ||
| + | |||
| + | CMV+EGFR-SigPep+mRuby3+StrepT+PolyA (F131+F152) -> P200, P201 | ||
| + | pASK_T7 RBS_LCN2_His (Vorlage für gewünschte Inserts) -> P202, P203 | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Concentrations: | ||
| + | P196: 385.3 ng/µl | ||
| + | P197: 462.6 ng/µl | ||
| + | P198: 84.8 ng/µl | ||
| + | P199: 96.0 ng/µl | ||
| + | |||
| + | *Send for sequencing: | ||
| + | **P201: FR12904454 (VF2) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Analytical digestion of P202 and P203 (pASK_T7 RBS_LCN2_His)=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment: ''' | ||
| + | Controll whether there is more than one NdeI restriction site. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Analytical digestion: | ||
| + | **The plasmid was digested with NdeI and HindIII | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |1 µl | ||
| + | |CutSmart buffer | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |NdeI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |HindIII | ||
| + | |- | ||
| + | |6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * Two bands were visible what means that there is only one NdeI restriction site. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | === Chemical biotinylation of BSA and eGFP=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 1 g BSA was chemically biotinylated with a 40x molar excess of biotin-NHS-ester: | ||
| + | |||
| + | * 100 ml of 50 mM borate buffer with 50 mM NaCl pH 8.85 | ||
| + | |||
| + | * dissolve BSA (10 mg/ml) | ||
| + | |||
| + | * add adequate amount of biotin-NHS-ester: 205 mg | ||
| + | |||
| + | * reaction for approx. 45 minutes at RT after full dissolving of biotin-NHS-ester | ||
| + | |||
| + | * store at 4 °C | ||
| + | |||
| + | eGFP was chemically biotinylated with a 16x (error in calculations) molar excess of biotin-NHS-ester: | ||
| + | |||
| + | * eGFP in 75 ml of 50 mM borate buffer without NaCl pH 8.85 | ||
| + | |||
| + | * 1to10 dilution: extinction(280) = 0.371 | ||
| + | |||
| + | * add adequate amount of biotin-NHS-ester: 69 mg | ||
| + | |||
| + | * reaction for approx. 45 minutes at RT after full dissolving of biotin-NHS-ester | ||
| + | |||
| + | * store at 4 °C | ||
| + | |||
| + | '''Next Steps''' | ||
| + | * add another 104 mg of biotin-NHS-ester incl. reaction at RT tp reach desired 40X molar excess | ||
| + | * Purification via column chromatography | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 12, August 1st - August 7th= | ||
| + | <div class="week" id="Week_12"> | ||
| + | |||
| + | =='''Monday, August 01'''== | ||
| + | |||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Cloning of Streptavidin/Streptactin/Traptactin from GSY5 and GSY6 into pASK75 + Transformation === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | *PCR products from GSY5 and GSY6 were, after digestion overnight with Nde1, loaded on an 1.8% agarose gel. The fragment bands at roughly 350 bps were extracted. | ||
| + | |||
| + | *Digested pASK75 (P202 and P203 dig. with Nde1) were loaded onto an 1% agarose gel, the vector fragments at 3800 bp isolated and purified. | ||
| + | |||
| + | *Vector fragment plus Streptavidin fragments were ligated and transformed into XL-1 blue | ||
| + | |||
| + | -> F159+F160, F159+F161 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Cloning of EGFR-TMD - mRuby - Strep with IRES-BirA=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | * 2 µg of P77 (EcoRI, XbaI) and P199 (EcoRI, SpeI) were digested for 2 hours at 37°C | ||
| + | * Digestions were separated on a 1% agarose gel, fragments corresponding to the vector (P77) and to the to be cloned insert (P199) were isolated and purified | ||
| + | * Ligation of inserts for 30 mins at RT | ||
| + | * Transformation of ligation into XL1-blue | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Retrafos of cell culture constructs for signal peptide testing for midipreps === | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | * Retransformation of all constructs in XL1-blue | ||
| + | * Constructs concerned are: P152, P171, P205 (Nanoluc-constructs) and P201, P208 and P211 (mRuby-constructs) | ||
| + | * Transformation according to protocol, plates incubated at 37°C overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, August 02'''== | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''Wednesday, August 03'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Verification of cloning P210 (CMV-IgKappa-Nanoluc-StrepTag-PolyA) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful cloning after sequencing showed that the last part of the construct might be missing | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * digestion with AgeI and PstI (sample was digested with EcoRI and PstI for 10 minutes, than inactivated an ligated) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16CMVIGKappaNanoluc.jpg|500px|]] | ||
| + | |||
| + | * the gel shows a band at about 3500 bp (pSB1C3 + full insert) | ||
| + | * gel shows additionally a band below 750 bp (CMV+ IgKappa) | ||
| + | * the sample contains the full insert as well as the small insert (CMV+IgKappa without Nanoluc-StrepTag-PolyA) | ||
| + | * cloning not succesful | ||
| + | * the construct was inocculated from plate again | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of ''E. coli'' XL1 blue with Ligations F159 + F160 and F159 + F161 (pASK + streptavidin from GSY 5 and 6)=== | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of ''E. coli'' XL1 blue. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator. | ||
| + | </div> | ||
| + | |||
| + | =='''Thursday, August 04'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | === MidiPrep P152 and P171 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' isolate Plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen). After washing the DNA with 70% Ethanol the pellet was left in special centrifugation tubes (labeled 152+QF; 171+QF) in order to air-dry. EB buffer for storage was later added by CG. | ||
| + | * Conc: 1000 ng/ul | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning of BAP Nanoluc (P206) before A3C5 (P213) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Preparative digestion: | ||
| + | |||
| + | P213 (A3C5): EcoRI and NgoMIV -> F170 | ||
| + | |||
| + | P206 (BAP Nanoluc): EcoRI and AgeI -> F171 | ||
| + | |||
| + | Both fragments were ligated after gelextraction. | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of Ligation was added to 100 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 750 µl LB-medium to each tube. | ||
| + | |||
| + | * Incubation for 1 hour at 37 °C | ||
| + | |||
| + | * The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker. | ||
| + | |||
| + | '''results:''' | ||
| + | Gelextraction: | ||
| + | [[File:Muc16BAPNanolucA3C5.jpg|500px]] | ||
| + | |||
| + | the plates showed no colonies. repeat | ||
| + | </div> | ||
| + | |||
| + | =='''Friday, August 05'''== | ||
| + | <div class="streptavidin"> | ||
| + | === Verification of successful cloning of all Streptavidin variants into pASK75 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Minipreps von pASK75 Streptavidin variants (codon optimzied Streptavidin, Streptactin, Traptactin) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MiniPrep was performed according to the manufacturer's protocol (QIAprep MiniPrep, Qiagen) for 3 clones of GSY5 and 6 clones of GSY6 | ||
| + | |||
| + | * Analytic digestion with 0.25 ''µ''L NdeI, 0.25 ''µ''L HindII, 1 ''µ''L SmartCut Buffer, 200 ng plasmid DNA, H<sub>2</sub>O to 20 µl | ||
| + | |||
| + | * 10 ''µ''L on 1% agarose gel electrophoresis | ||
| + | |||
| + | '''Results:''' Successful cloning was verified for all plasmids by analytical digestion. Expected and observed fragment size for the vector was 3.1 kb and about 350 bp for the inserts. 4 plasmids were sent for sequencing (1x GSY5, 3x GSY6). Alle sequences were error-free. Therefore all 3 Streptavidin variants are available now. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Transformation of: Streptavidin variants, PAS-Plasmid, Interlab-Plasmids, Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA and Ligation of BAP-Nanoluc-A3C5=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation of Streptavidin variants, PAS-Plasmid, Interlab-Plasmids into E. coli BL21 and Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA, Ligation of BAP-Nanoluc-A3C5 in E. coli XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of Ligations mixes were added to 50 µl of competent cells and gently mixed, 1 µl for ccc Plasmids | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 1 min. heat shock at 42 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube, and incubation for 1 h at 37°C | ||
| + | |||
| + | * The cell suspension was plated on ampicillin plates (inclusive rescue plate) / CamR- plates and incubated for 7 h at 37 °C in the programmed incubator. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA and Ligation of BAP-Nanoluc-A3C5=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Re-Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA, Re-Ligation of BAP-Nanoluc-A3C5 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | The corresponding Fragments were mixed with 2 µl Ligase-buffer, 1 µl T4-Ligase and filled with water up to 20 µl | ||
| + | |||
| + | * 7.3 µl F163 + 2.7 µl F162 (=F173) | ||
| + | |||
| + | * 3.7 µl F170 + 6.3 µl F171 (=F174) | ||
| + | |||
| + | The Ligation mixes were incubated for 1 h at room temperature. | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === SDS Page of Streptavidin production === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful Streptavidin production and preperation for the data package | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Streptavidin samples from the production process were further processed with SDS buffer. Some samples had very few volume, therefore the overall volume was reduced. For informations on the samples content please refer to NA. The low volume might be a problem due to evaporation issues. | ||
| + | |||
| + | * Sample 1: the tube was empty | ||
| + | * Sample 2: 1 µl sample was mixed with 7 µl water and 2 µl SDS buffer | ||
| + | * Sample 3: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | * Sample 4: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | * Sample 5: 20 µl sample was mixed with 5 µl SDS buffer | ||
| + | * Sample 6: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | * Sample 7: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | * Sample 8: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | * Sample 9: 80 µl sample was mixed with 20 µl SDS buffer | ||
| + | |||
| + | 10 µl of each sample were loaded on the pre-mixed SDS gel. | ||
| + | |||
| + | Gel 1: 8 µl Ladder, samples 1-6 | ||
| + | |||
| + | Gel 2: 8 µl Ladder, samples 7-9, 5 µl of samples 7-9 | ||
| + | |||
| + | The gel ran at 160 V for about 1 1/2 h. Afterwards it was incubated for 30 min. in staining solution, 10 min. in destaining solution 1 and over the weekend in destaining solution 2. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16SDSstrep2.jpg|500px]] | ||
| + | |||
| + | [[File:Muc16SDSstrep.jpg|500px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 13, August 8th - August 14th= | ||
| + | <div class="week" id="Week_13"> | ||
| + | |||
| + | =='''Monday, August 08'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === PCR, Purification and digestion of BirA === | ||
| + | |||
| + | '''Investigator: LK ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of BirA | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR according to the SOP (primers: O17 and O18, Annealing at 62°C) | ||
| + | * PCR purification via the purification kit according to the manufacturer's protocol | ||
| + | * preparative digestion with XbaI and PstI over night in a 50µl batch | ||
| + | |||
| + | '''Results:'''' | ||
| + | * Concentration after PCR-purification: 116ng/µl | ||
| + | * Gel: One thick band at ~1000Bp --> BirA and one at ~2100Bp --> vector, we're cut out and purified via the Gelextraction kit according to the manufacturer's protocoll. | ||
| + | * Concentrations: BirA (F186) : 62,5 ng/µl, pSB1C3 (F187): 32,5 ng/µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Determination of Streptavidin concentration === | ||
| + | |||
| + | '''Investigator: NA ''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * OD280 measurement with quartz cuvette | ||
| + | |||
| + | '''result:''' the 1:100 dilution showed an OD of 0,469 | ||
| + | |||
| + | Calculation with extinction coefficient (expasy tool) of 41940 M <sup>-1</sup> * cm <sup>-1</sup> and 13.3 kDA as molar mass of streptavidin monomer provides concentration below: | ||
| + | |||
| + | 14.87 mg/mL | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inocculation of Streptavidin (wild type, P235) in M9 medium for fermentation === | ||
| + | |||
| + | '''Investigator: NA ''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * M9 medium with Amp (1:1000) is inocculated with preculture (~50 mL) of P235 | ||
| + | |||
| + | * the culture is incubated in the shaker over night at 37°C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === PCR purification === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Purificate bands for blunt ligation in F113 | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Seeding of HEK293T cells in cell culture=== | ||
| + | |||
| + | '''Investigator: JB''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Thawing and seeeding of HEK293T cells for experiments later this week | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Tube with cells was thawed quickly in a 37°C waterbath | ||
| + | |||
| + | * After bringing most of the cell suspension in a fluid state, the suspension was transferred into a 50 ml Falcon tube and suspended in 50 ml of RPMI1640 medium (with 10% CFS and Pen/Strep added). | ||
| + | |||
| + | * Cells were spun down for 2 mins at 2,300 rcf and the pellet resuspended in 20 ml of medium in order to remove residual DSMO from the cells (DSMO having been used for freezing the cells). | ||
| + | |||
| + | * The resuspended cells were transferred into a culture flask and incubated overnight at 37°C/5% CO². | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Cloning of IgKappa signale peptide, PAS-Lysin and further constructs for the receptor=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Cloning of CMV-IgKappa+Nanoluc-Strep-PolyA, BAP-Nanoluc+A3C5, IRES-BirA + polyA and Sumo-PAS-Lysin into pASK75 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Digestion of 3 µg of each plasmid in 30 µl mix: | ||
| + | * 20 µl P205 (CMV-IgKappa) with EcoRI, AgeI -> Gel 1, left lane, expected fragment at about 600 bp -> F175 | ||
| + | * 15 µl P160 (Nanoluc-Strep-PolyA) with EcoRI, NgoMiV -> Gel 1, right lane, expected fragment at about 3000 bp -> F176 | ||
| + | * 20 µl P212 (A3C5) with EcoRI, NgoMiV -> Gel 2, left lane, expected fragment at about 2.200 bp -> F177 | ||
| + | * 10 µl P206 (BAP-Nanoluc) with EcoRI, AgeI -> Gel 2, right lane, expected fragment at about 500 bp -> F178 | ||
| + | * 10 µl P199 (EGFR-TMD-mRuby-Strep) with EcoRI, SpeI -> Gel 3, left lane, expected fragment at about 1.500 bp -> F179 | ||
| + | * 20 µl P236 (PAS-Lysin-Sumo) with NdeI, HindIII -> Gel 3, right lane, expected fragment at about 1.000 bp -> F180 | ||
| + | * 10 µl P77 (IRES-BirA-KDEL) with EcoRI, SpeI -> Gel 4, left lane, expected fragment at about 1000 bp -> F181 | ||
| + | |||
| + | Ligation in 20 µl mix (s. above fragment names): | ||
| + | * F182: 2.9 µl F176 + 7.1 µl F175 | ||
| + | * F183: 5.2 F177 + 4.8. F178 | ||
| + | * F184: 7.0 µl F159 + 3.0 µl F180 | ||
| + | * F185: 2.9 µl F133 + 7.1 µl F181 | ||
| + | |||
| + | Transformation according to SOP. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | For all ligations colonies grew the next day. | ||
| + | |||
| + | Gel 1: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160808_1.jpg|500px]] | ||
| + | |||
| + | Gel 2: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160808_2.jpg|500px]] | ||
| + | |||
| + | Gel 3: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160808_3.jpg|500px]] | ||
| + | |||
| + | Gel 4: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160808_4.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, August 09'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Transformation of Biobricks in NEB Turbo === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Acquisition of pBAD/AraC and Lox site | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * resuspend DNA with 10 µl ddH2O in well of interest (standard distribution kit) | ||
| + | * incubate for 5 min | ||
| + | * transformation of 1 µl plasmid according to SOP in NEB Turbo | ||
| + | |||
| + | Used bricks: | ||
| + | BBa_I0500 and BBa_J61046 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation and induction of AraC+pBAD controlled eGFP production in E.coli BL-21 === | ||
| + | |||
| + | '''Investigator: LK ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Confirmation of arabinose dependend activation of the pBAD-promotor in AraC-Operon. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * E.coli Bl-21 with P61 (PC+AraC+pBAD+eGFP in pSB1C3) we're picked and a 4ml preculture (LB+cam) was inoculated and incubated for 6h. | ||
| + | * According to the following table 50ml cultures (LB+cam) we're incoculated with 500µl of the preculture and incubated over night at 37°C. | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Arabinose concentration:''' | ||
| + | |negative control | ||
| + | |100mM | ||
| + | |10mM | ||
| + | |1mM | ||
| + | |100µM | ||
| + | |10µM | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of F186 (BirA) into F187 (pSB1C3) === | ||
| + | |||
| + | '''Investigator: LK ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Getting the BirA for the prokaryotic receptor. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1,7µl of F187 and 6,3µl of F186 we're ligated for 1h at RT accoridng to the SOP. --> F189 | ||
| + | * The ligation-batch was transformed into NEB-Turbo cells and plated out on cam-plates. Incubation at 37° o/n. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Inoculation of precultures from Ligations F182-F185 === | ||
| + | |||
| + | '''Investigator: LK ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Getting plamids of CMV-IgKappa+Nanoluc-Strep-PolyA (F182), BAP-Nanoluc+A3C5 (F183),Sumo-PAS-Lysin (F184) and IRES-BirA + polyA (F185) | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 4mL cultures (LB cam resp. amp (F184)). Incubation at 37°C o/n. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | === Inocculation of fermenter === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * centrifuge preculture (P235 in M9 medium) | ||
| + | * resuspend pellet in HCDF medium and inocculate fermenter (HCDF medium) | ||
| + | * pO<sub>2</sub>= 60 %; Temp= 30°C, pH~7 | ||
| + | * growth over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === SDS Page of Streptavidin production === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful Streptavidin production and preperation for the data package | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Streptavidin samples from the production process were further processed with SDS buffer. Some samples had very few volume, therefore the overall volume was reduced. | ||
| + | |||
| + | * 1: 8 µl Ladder | ||
| + | * 2: 10 µl after induction (1 µl sample was mixed with 7 µl water and 2 µl SDS buffer) | ||
| + | * 3: 5 µl supernantant after denaturation (20 µl sample was mixed with 20 µl H<sub>2</sub>O and 10 µl SDS buffer) | ||
| + | * 4: 5 µl before precipitation(80 µl sample was mixed with 20 µl SDS buffer) | ||
| + | * 5: 10 µl supernantant after 40% precipitation (80 µl sample was mixed with 20 µl SDS buffer) | ||
| + | * 6: 10 µl supernantant after 70% precipitation (80 µl sample was mixed with 20 µl SDS buffer) | ||
| + | * 7: 10 µl after dissolving in 2.2M (NH<sub>4</sub>)2SO<sub>4</sub> (80 µl sample was mixed with 20 µl SDS buffer) | ||
| + | |||
| + | |||
| + | The gel ran at 145 V | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16sa31.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Transformation of F192 and P238 === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl2 competent ''E. coli'' XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice. | ||
| + | |||
| + | * 7 µl of DNA was added to 50 µl of competent cells and gently mixed. | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min. heat shock at 37 °C | ||
| + | |||
| + | * Adding of 950 µl LB-medium to each tube. | ||
| + | |||
| + | * The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C. | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, August 10'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Minipreps and analytical digestion of P237 - P243 === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Isolating and verifiing plamids of CMV-IgKappa+Nanoluc-Strep-PolyA (F182), BAP-Nanoluc+A3C5 (F183),Sumo-PAS-Lysin (F184) and IRES-BirA + polyA (F185) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Minipreps were performed according to the manufacturer's protocol. | ||
| + | * ~200- 500ng of ach Plasmid (accept P241) were digested with EcoI and PstI for 45 min. and a gel-elctrophoresis (1% Agarose) of the digestion-batches was performed. | ||
| + | |||
| + | '''Results:''' | ||
| + | * P237: expected band for CMV-IgKappa+Nanoluc-Strep-PolyA at ~1850 Bp , but unexpected band at ~1100. Discard sample? | ||
| + | * P238: expected band for CMV-IgKappa+Nanoluc-Strep-PolyA at ~1850 Bp | ||
| + | * P239 and P240: expected band at ~650 Bp | ||
| + | * P242 and P243: bands at ~2000 Bp. Digested vector and Fragment are not distinguishable --> sequencing | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Cloning constructs for the eukaryotic and prokaryotic receptor=== | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Cloning of EGFR-TMD-mRuby-Strep-IRES-BirA-KDEL-PolyA, BAP-Nanoluc-A3C5-EspP-2xTerm | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Digestion of 3 µg of each plasmid in 30 µl mix: | ||
| + | * 6 µl P242 (IRES-BirA-KDEL-PolyA) with EcoRI, XbaI -> Gel 1, left lane, expected fragment at about 4000 bp -> F190 | ||
| + | * 20 µl P108 (EspP-Termin.) with EcoRI, NgoMiV -> Gel 1, right lane, expected fragment at about 3000 bp -> F191 | ||
| + | |||
| + | Ligation in 20 µl mix (s. fragment names above): | ||
| + | * F192: 4 µl F190 + 6 µl F179 | ||
| + | |||
| + | |||
| + | Transformation according to SOP. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | Gel 1: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160810_1.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Transformation of P22; P67; P122; P137; P160 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of P22; P67; P122; P137; P160 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |P22 | ||
| + | |K157001 | ||
| + | |RFP-Gen.(Retrafo P4) | ||
| + | |- | ||
| + | |P67 | ||
| + | |pSB1C3 | ||
| + | |EGFR signal peptide (transloc. pore) | ||
| + | |- | ||
| + | |P122 | ||
| + | |pSB1C3 | ||
| + | |Ig Kappa (digest. GSY3) | ||
| + | |- | ||
| + | |P137 | ||
| + | |pSB1C3 | ||
| + | |T7/BM40 aus GSY | ||
| + | |- | ||
| + | |P160 | ||
| + | |pSB1C3 | ||
| + | |NanoLuc + StrepTag + Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Fermentation === | ||
| + | |||
| + | '''Investigator:''' NA | ||
| + | |||
| + | '''Aim of the experiment:''' Streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * exponential feed (glucose, MgSO<sub>4</sub>, Thiamin, Amp, HCDF salt, micronutrients) for ~13 h (about 3-4 L) | ||
| + | * Induction of streptavidin expression (anhydro tetracyclin) at OD= 111.2 | ||
| + | * production over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, August 11'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Minipreps and analytical digestion of BBa_I0500 (AraC+ pBAD), BBa_J61046 (LoxP) and F189 (BirA) === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Isolating and verifiing the plasmids. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The Preperation was performed according to the manufacturer's protocol. | ||
| + | * Each plasmid was digested with EcoRI and PstI and a gel-elctrophoresis (1% Agarose) of the digestion-batches was performed. | ||
| + | |||
| + | '''Results:''' | ||
| + | * BBa_I0500 (AraC+ pBAD) = P246 and P247: Band at ~1250; Cloning was succesful | ||
| + | * BBa_J61046 (LoxP) = P244 and P245: Fragment is too small (34Bp), not visible on gel. | ||
| + | * F189 (BirA) = P248: Band at ~ 1000Bp; Cloning was succesful | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Cloning of Streptavidin-variants into pASK111=== | ||
| + | |||
| + | '''Investigator: JB, LK, CG''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Cloning of Streptactin, Traptavidin, Streptavidin into CamR pASK111 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Digestion of about 2.5 µg of each plasmid in 30 µl mix: | ||
| + | * 25 µl P233 (Streptavidin) with XbaI, HindIII -> Gel 1, left lane, expected fragment at about 400 bp -> F195 | ||
| + | * 25 µl P231 (Traptavidin) with XbaI, HindIII -> Gel 1, right lane, expected fragment at about 400 bp -> F196 | ||
| + | * 15 µl P241 (PAS-Lysin) with XbaI, HindIII -> Gel 2, left lane, expected fragment at about 400 bp -> - | ||
| + | * 25 µl P229 (Streptactin) with XbaI, HindIII -> Gel 2, left right, expected fragment at about 400 bp -> F197 | ||
| + | * 15 µl P249 (pASK111) with XbaI, HindIII -> Gel 3, left lane, expected fragment at about 3000 bp -> F198 | ||
| + | |||
| + | Ligation in 20 µl mix overnight (s. fragment names above): | ||
| + | * F199: 3.7 µl F195 + 6.3 µl F198 | ||
| + | * F200: 5.4 µl F196 + 4.6 µl F198 | ||
| + | * F201: 4.1 µl F197+ 5.9 µl F198 | ||
| + | |||
| + | |||
| + | Transformation according to SOP. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | Gel 1: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160811_1.jpeg|500px]] | ||
| + | |||
| + | Gel 2: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160811_2.jpg|500px]] | ||
| + | |||
| + | Gel 3: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160811_3.jpg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="polymerization"> | ||
| + | |||
| + | ===Testpolymerization: BSA-coating=== | ||
| + | |||
| + | '''Investigator: EU, JH''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Coating of 6-well plate with BSA according to a modified ELISA-protocol. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Bicarbonate/carbonate coating buffer (100mM): | ||
| + | 30,3 mg Na2CO3 | ||
| + | 60 mg NaHCO3 | ||
| + | 10 ml distilled water | ||
| + | |||
| + | "Blocking" Buffer: | ||
| + | 1% BSA in PBS | ||
| + | |||
| + | Each well was covered with 0,5 ml BSA buffer and 0,5 ml bicarbonate buffer. After overnight incubation at RT, solution in wells was aspirated one by one and streptavidin was added. After a short incubation, the streptavidin(&Tris/HCl) solution was aspirated and Biotin solution was added (by needle). | ||
| + | |||
| + | 1. well: 200 µl Streptavidin (11,2 mg/ml) + 200 µl Biotin, 2. well: 400 µl Streptavidin and 400 µl Tris/HCl + 200 µl Biotin, 3. well: 500 µl Streptavidin and 500 µl Tris/HCl + 300 µl Biotin, 4. well: 500 µl Streptavidin + 10 µl Biotin (this time with 2,5 µl pipette) | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Overall polymerization was observed again, although it seemed more like an aggregation than a polymerization. | ||
| + | No coating was observed except in the 4. well. Biotin-injection speed may be the critical point. | ||
| + | |||
| + | Poly-D-Lysine coating will follow as next step. | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Transformation of P64; P73 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of P22; P67; P122; P137; P160 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |P64 | ||
| + | |pSB1C3 | ||
| + | |VEGF-p2a-PDGF | ||
| + | |- | ||
| + | |P73 | ||
| + | |pSB1C3 | ||
| + | |Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Over night liquid culture of P22; P67; P122; P137; P160 === | ||
| + | |||
| + | '''Investigator: EF, MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === harvesting streptavidin === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * OD= 174 | ||
| + | * centrifuge culture | ||
| + | * freeze pellet | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === SDS PAGE Streptavidin fermentation === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successfull production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * according to protocol | ||
| + | * 8 µl Ladder; before induction; after induction | ||
| + | * both samples: 3 µl culture + 77 µl TB + 20 µl SDS buffer; 12 µl on SDS gel | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16FermentationVORNACH2.1.jpg|500px]] | ||
| + | |||
| + | *The fermentation was not successful, streptavidin band (unter 15 kDa marker) is just a bit thicker. The reason might be the ampR of the transformed plasmid. Due to the high cell concentration beta lactamase (secreted in periplasma) gets in the medium and hydrolyses amp. As a result cells lose plasmid because there is no selective pressure without an antibiotic in medium. | ||
| + | |||
| + | * next step: cloning pf streptavidin mutants into pASK111 (camR) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Sequencing of P238, P245, P248 === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) - all samples: VF2 | ||
| + | * barcodes in stock list | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, August 12'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === PCR of Gensythesis (GSY) 7 and 8 === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of GSY 7 and 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''25 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |5X Q5 Reaction Buffer | ||
| + | |5 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 mM dNTPs | ||
| + | |0.5 µL | ||
| + | |200 μM | ||
| + | |- | ||
| + | |10 μM Forward Primer (D20) | ||
| + | |1.25 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (VR) | ||
| + | |1.25 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Q5 High-Fidelity DNA Polymerase | ||
| + | |0.25 μl | ||
| + | |0.02 U/μl | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 25 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||5 - 10 sec | ||
| + | |- | ||
| + | |61 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |16 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Note: Use of the NEB T m Calculator is highly recommended. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | After running on 1 %agarose gel only the 2kb bands belonging to GSY8 could be distinguished. These were extracted from the gel. | ||
| + | |||
| + | |||
| + | -> Repeat PCR for GSY7 | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Gelextraction of GSY8 === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' Gelextraction of 2kb band of PCR belonging to GSY8 | ||
| + | '''Procedure:''' | ||
| + | |||
| + | Gelextraction was performed by manufacturers protocol (SLG). | ||
| + | |||
| + | '''Concentration:''' | ||
| + | |||
| + | 7,7 ng/ul | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | |||
| + | |||
| + | === Plasmid preparation of liquid culture of P22; P67; P122; P137; P160 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of P22; P67; P122; P137; P160 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |P22 | ||
| + | |K157001 | ||
| + | |RFP-Gen.(Retrafo P4) | ||
| + | |- | ||
| + | |P67 | ||
| + | |pSB1C3 | ||
| + | |EGFR signal peptide (transloc. pore) | ||
| + | |- | ||
| + | |P122 | ||
| + | |pSB1C3 | ||
| + | |Ig Kappa (digest. GSY3) | ||
| + | |- | ||
| + | |P137 | ||
| + | |pSB1C3 | ||
| + | |T7/BM40 aus GSY | ||
| + | |- | ||
| + | |P160 | ||
| + | |pSB1C3 | ||
| + | |NanoLuc + StrepTag + Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * According to Mini-prep kit, SLG | ||
| + | |||
| + | '''Results:''' | ||
| + | * P22 ->P1001 103.6 ng/µL | ||
| + | * P67 ->P1004 92.9 µL | ||
| + | * P122 -> P1000 80.8 ng/µL | ||
| + | * P137 -> P1003 87.9 µL | ||
| + | * P160 -> P1002 100 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Dephosphorylation of F113[HincII digested] === | ||
| + | |||
| + | '''Investigator: MP, EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Dephosphorylation of F113[HincII digested] (RFP-Reportercassette ) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | According to NEB online Protocol for Dephosphorylation of 5 ́-ends of | ||
| + | DNA using Antarctic Phosphatase (M0289): | ||
| + | * 12 µL F113 (85.1 ng/µL) | ||
| + | * 2 µL Antarctic Phosphatase Rxn Buffer (10X) | ||
| + | * 1 µL Antarctic Phosphatase (5000 u/mL) | ||
| + | * 5 µL ddH2O | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Midiprep P238 (CMV-IgKappa-Nanoluc-Strep-polyA) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | P253 - concentration: 2134 ng/µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion, Gel extraction and ligation of F1000 (P122), F1001 (P67), F1002 (P160) and F1003 (P137) === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Preparation of plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 33 µL DNA | ||
| + | * 1 µL enzyme 1 | ||
| + | * 1 µL enzyme 2 | ||
| + | * 5 µL cut smart buffer | ||
| + | * 10 µL ddH20 | ||
| + | |||
| + | Enzymes used: | ||
| + | * F1000: AgeI + PstI | ||
| + | * F1001: AgeI + PstI | ||
| + | * F1002: NgoMiT + PstI | ||
| + | * F1003: AgeI + PstI | ||
| + | |||
| + | Gelextraction: | ||
| + | * Using gelextraction kit, Quiagen | ||
| + | |||
| + | Ligation | ||
| + | * 2µl F1000 + 6µl F1002 | ||
| + | * 2µl F1001 + 6µl F1002 | ||
| + | * 2µl F1003 + 6µl F1002 | ||
| + | + 1µl T4 Ligase and 1 l T4 ligase buffer for each ligation | ||
| + | |||
| + | '''Results:'''' | ||
| + | |||
| + | Left to right F1002(2x), F1001(2x), F1000(2x) and F1003(1x) | ||
| + | |||
| + | * F1000 30 ng/µL | ||
| + | * F1001 23,8 ng/µL | ||
| + | * F1002 9,4 ng/µL | ||
| + | * F1003 23,1 ng/µL | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Saturday, August 13'''== | ||
| + | |||
| + | <div class="linkers"> | ||
| + | |||
| + | === Miniprep of pASK111 (empty) and diegestion === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of pASK111 and digestion for further ligation with SUMO-PAS-Lysin. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The Miniprep was performed according to the manufacturer's protocol. | ||
| + | * The whole DNA was diegested with XbaI and HindIII. Over night at 37°C. | ||
| + | * Gelelectrophoresis was performed and showed a significant band at ~2700. | ||
| + | |||
| + | '''Comment''' | ||
| + | * Gel-ectraction has yet to be done . | ||
| + | * equally digested insert from P236 was visible on gel. --> no ligation possible. | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F1006; F1007; F1008 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of F1006; F1007; F1008 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Ligation''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1006 | ||
| + | |F1000 + F1002 | ||
| + | |pSB1C3 | ||
| + | |Ig Kappa + NanoLuc + StrepTag + Poly-A | ||
| + | |- | ||
| + | |F1007 | ||
| + | |F1003 + F1002 | ||
| + | |pSB1C3 | ||
| + | |T7/BM40 + NanoLuc + StrepTag + Poly-A | ||
| + | |- | ||
| + | |F1008 | ||
| + | |F1001 + F1002 | ||
| + | |pSB1C3 | ||
| + | |EGFR + NanoLuc + StrepTag + Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion and gelextraction of P64; P73 === | ||
| + | |||
| + | '''Investigator: JB, MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Preparation for ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL DNA | ||
| + | * 1 µL enzyme 1 | ||
| + | * 1 µL enzyme 2 | ||
| + | * 2 µL buffer | ||
| + | * 6 µL ddH20 | ||
| + | |||
| + | Enzymes used: | ||
| + | * P64: EcoRI + SpeI | ||
| + | * P73: EcoRI + XbaI | ||
| + | |||
| + | Gelextraction: | ||
| + | * Using gelextraction kit, Quiagen | ||
| + | |||
| + | '''Results:'''' | ||
| + | |||
| + | [[File:Muc16_digestion_P64_P73.png|250px]] | ||
| + | |||
| + | Left: P64 Right: P73 | ||
| + | |||
| + | * P64 -> F1011 14,2 ng/µL | ||
| + | * P73 -> F1012 17,6 ng/µL | ||
| + | </div> | ||
| + | |||
| + | =='''Sunday, August 14'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ===Miniprep of P199-P201 and analytical digestion === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Isolation of "Streptavidin-variants" in pASK111 for preinoculation tomorrow. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The Miniprep was performed according to the manufacturer's protocol. | ||
| + | * ~200ng where digested with XbaI and HindIII. | ||
| + | |||
| + | '''Results:''' | ||
| + | * All samples showed significant bands at ~400 Bp. | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Transformation of P199-201 into E.coli BL-21=== | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Introducing Strep-variants in pASK111 into BL-21 for further fermentation. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The cells were transformed according to the SOP with 1µl of DNA and after 1h of incubation in LB without antibiotics plated out on Cam-plates. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ===Miniprep of P113-P213 and analytical digestion === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | '''Aim of the experiment:''' Isolation of "Streptavidin-variants" in pASK111 for preinoculation tomorrow. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The Miniprep was performed according to the manufacturer's protocol. | ||
| + | * ~200 ng were digested with EcoRI and PstI. | ||
| + | |||
| + | '''Results:''' | ||
| + | * | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === PCR of Gensynthesis (GSY) 7, 8 === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of GSY7 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) | ||
| + | * Different amounts of template DNA were used | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |5X Q5 Reaction Buffer | ||
| + | |10 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 mM dNTPs | ||
| + | |1 µL | ||
| + | |200 μM | ||
| + | |- | ||
| + | |10 μM Forward Primer (D20) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (VR) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |1 - 10 ng | ||
| + | |- | ||
| + | |Q5 High-Fidelity DNA Polymerase | ||
| + | |0.5 μl | ||
| + | |0.02 U/μl | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|40 cycles||98 °C||5 - 10 sec | ||
| + | |- | ||
| + | |58 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | *PCR GSY8 successful. | ||
| + | |||
| + | -> Do it again for GSY 7! | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Over night liquid culture of F1006; F1007; F1008 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Gelextaction of F1004 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Using gelextraction kit, Quiagen | ||
| + | |||
| + | '''Results:'''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_F1004.png|250px]] | ||
| + | |||
| + | * long F1004 -> F1013 6,4 ng/µL | ||
| + | * short F1004 -> F1014 1,1 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation F1012 + F1011 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Ligation F1012 + F1011 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmind''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1011 | ||
| + | | | ||
| + | |VEGF-p2a-PDGF | ||
| + | |- | ||
| + | |F1012 | ||
| + | |pSB1C3 | ||
| + | |Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL F1012 | ||
| + | * 6 µL F1011 | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 Ligase | ||
| + | * 8 µL ddH2O | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1015 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F1015 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of F1015 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Ligation''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1015 | ||
| + | |F1011 + F1012 | ||
| + | |pSB1C3 | ||
| + | |VEGF-p2a-PDGF + poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | |||
| + | '''Results:''' | ||
| + | * No colonies. Do it again. | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--- this closes the week --> | ||
| + | </div> | ||
| + | <!--- ^^^^ this closes the week --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | <!--- PLEASE DO NOT TOUCH !!!! --> | ||
| + | |||
| + | =Week 14, Aug 15th - Aug 21st= | ||
| + | |||
| + | <div class="week" id="Week_14"> | ||
| + | |||
| + | =='''Monday, Aug 15th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion of P251 and P252 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verify cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 1µL Plasmid was digested with 1µL of EcoRI and PstI | ||
| + | * Digestions were incubated for 1h | ||
| + | * Samples were seperated by gelelectrophoresis on a 1% agarose gel | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Cloning seems to have worked | ||
| + | |||
| + | |||
| + | [[File:Muc16_P251_P252.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Plasmidpreparation of liquid culture of F1006; F1007; F1008 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of F1006; F1007; F1008 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1006 | ||
| + | |pSB1C3 | ||
| + | |Ig Kappa + NanoLuc + StrepTag + Poly-A | ||
| + | |- | ||
| + | |F1007 | ||
| + | |pSB1C3 | ||
| + | |T7/BM40 + NanoLuc + StrepTag + Poly-A | ||
| + | |- | ||
| + | |F1008 | ||
| + | |pSB1C3 | ||
| + | |EGFR + NanoLuc + StrepTag + Poly-A | ||
| + | |} | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * According to Mini-prep. kit, Quiagen | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1006 -> F1005 83.2 ng/µL | ||
| + | * F1007 -> F1006 91.8 ng/µL | ||
| + | * F1008 -> F1007 367.1 ng/µL | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Cloning of Nanoluc-Strep in front of double terminator and PAS-Lysine into pASK111=== | ||
| + | |||
| + | '''Investigator: LK, CG''' | ||
| + | |||
| + | '''Aim of experiment:''' | ||
| + | |||
| + | Cloning of Nanoluc-Strep in front of double terminator and PAS-Lysine into pASK111 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Plasmid preperation according to SOP of P264, P265 | ||
| + | |||
| + | Digestion of about 3 µg of each plasmid in 30 µl mix: | ||
| + | * 6 µl P29 (double teminator) with EcoRI, XbaI -> Gel 1, left lane, expected fragment at about 3000 bp -> F207 | ||
| + | * 15 µl P98 (Nanoluc-Strep) with EcoRI, SpeI -> Gel 1, right lane, expected fragment at about 500 bp -> F208 | ||
| + | * 15 µl P264 (PAS-Lysin) with XbaI, HindIII -> Gel 2, left lane, expected fragment at about 1000 bp -> F210 | ||
| + | |||
| + | Ligation in 20 µl mix for 30 min. at room temperature (s. fragment names above): | ||
| + | * F209: 3.7 µl F207 + 6.3 µl F208 | ||
| + | * F211: 1.1 µl F210 + 8.9 µl F206 | ||
| + | |||
| + | |||
| + | Transformation according to SOP. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | Gel 1: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160816_1.jpeg|500px]] | ||
| + | |||
| + | Gel 2: | ||
| + | |||
| + | [[File:Muc16_PrepDigest_160816_2.jpeg|500px]] | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, Aug 16th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Over night liquid culture of P64; P73 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Gelextraction of PCR GSY8(Saturday, August 13) === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of PCR GSY8(Sunday, August 14) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Using gelextraction kit, SLG | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1016 3,4 ng/µL | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Seeding of HEK293T cells for transfection === | ||
| + | |||
| + | '''Investigator: LK, CG ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | 3x4 petri dishes with HEK293T cells for performing the Nanoluc assay in order to validate the signale peptides | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Washing each T75 flask with 10 ml PBS | ||
| + | * Centrifugation for 10 min. at 1.8 rcf | ||
| + | * Resuspending the cell pellet in 10 ml medium | ||
| + | * Counting the cells (33x10^6 cells in 10 ml) | ||
| + | * Plating 0.5x10^6 cells onto petri dishes in 10 ml medium an 1x10^6 onto a T75 flask | ||
| + | * Incubation for 2 days | ||
| + | |||
| + | '''Results:''' | ||
| + | * ready to transfect cells in petri dishes | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Gelextraction of PCR GSY8(Saturday, August 13) === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of PCR GSY8(Sunday, August 14) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Using gelextraction kit, SLG | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1016 3,4 ng/µL | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F1015 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of F1015 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Ligation''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1015 | ||
| + | |F1011 + F1012 | ||
| + | |pSB1C3 | ||
| + | |VEGF-p2a-PDGF + poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === PCR of Gensynthesis (GSY) 7 === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of GSY7 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) | ||
| + | * Reactions with and without GC Enhancer were performed | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction x 6''' | ||
| + | |'''Final concentration''' | ||
| + | |-*Product could not be purified | ||
| + | |5X Q5 Reaction Buffer | ||
| + | |10 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 mM dNTPs | ||
| + | |1 µL | ||
| + | |200 μM | ||
| + | |- | ||
| + | |10 μM Forward Primer (D20) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (VR) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |1 ng | ||
| + | |- | ||
| + | |Q5 High-Fidelity DNA Polymerase | ||
| + | |0.5 μl | ||
| + | |0.02 U/μl | ||
| + | |- | ||
| + | |5X Q5 High GC Enhancer (optional) | ||
| + | |5 µL | ||
| + | |1X | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|40 cycles||98 °C||5 - 10 sec | ||
| + | |- | ||
| + | |58 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |16 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | *negativ. Two 1% agarose gels were performed to prove the PCR product. Both were negativ. | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Precipitation of eGFP === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of experiment:''' Determination of (NH<sub>4</sub>)<sub>2</sub>SO<sub>4</sub> -concentration needed to precipitate eGFP. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 50mL of eGFP solution (from fermentation) where gradually precipitated with rising concentrations (NH<sub>4</sub>)<sub>2</sub>SO<sub>4</sub> (step size: 400mM) | ||
| + | * All steps where performed at 4°C. | ||
| + | * Pellets where solved in 10 mL 1x PBS. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | ''Extinction:'' | ||
| + | * Maximum Absorption at 488nm for eGFP. Absorption coefficient(eGFP,488): 53000 mol/Lcm. | ||
| + | * | ||
| + | {|cellspacing="1" border="1" | ||
| + | |'''mM (NH<sub>4</sub>)<sub>2</sub>SO<sub>4</sub>''' | ||
| + | |'''Dilution''' | ||
| + | |'''Extinction''' | ||
| + | |'''Concentration eGFP [µM]''' | ||
| + | |- | ||
| + | |400 | ||
| + | |1:10 | ||
| + | |0,197 | ||
| + | |37,0 | ||
| + | |- | ||
| + | |800 | ||
| + | |1:10 | ||
| + | |0,084 | ||
| + | |15,8 | ||
| + | |- | ||
| + | |1200 | ||
| + | |1:10 | ||
| + | |0,457 | ||
| + | |86,2 | ||
| + | |- | ||
| + | |1600 | ||
| + | |1:100 | ||
| + | |0,16 | ||
| + | |301,8 | ||
| + | |- | ||
| + | |2000 | ||
| + | |1:100 | ||
| + | |0,375 | ||
| + | |707,5 | ||
| + | |- | ||
| + | |2400 | ||
| + | |1:10 | ||
| + | |0,505 | ||
| + | |95,3 | ||
| + | |- | ||
| + | |2800 | ||
| + | |1:10 | ||
| + | |0,08 | ||
| + | |15,0 | ||
| + | |} | ||
| + | |||
| + | [[File:Muc16_sds-page_160818.jpeg|250px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inocculation of P258_E.coli_Bl-21 in M9 medium for fermentation === | ||
| + | |||
| + | '''Investigator: LK ''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * M9 medium with Cam (1:1000) was inocculated with preculture (~50 mL) of P258 (pASK111+Streptavidin) | ||
| + | |||
| + | * the culture was incubated in the shaker over night at 37°C | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of F1005; F1016 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Phosphorylation of F1005; F1016 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 39 µL DNA | ||
| + | * 5 µL 10x T4 PNK Bufer | ||
| + | * 10 mM ATP | ||
| + | * T4 PNK | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Phosphorylation of O31; O32 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Phosphorylation of O31; O32 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 0.5 µL Oligonucleotides (10µM) | ||
| + | * 2 µL 10x T4 PNK Bufer | ||
| + | * 2 µL 10 mM ATP | ||
| + | * 1 µL T4 PNK | ||
| + | * 14.5 µL H2O | ||
| + | * 95 °C; 3 min | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Hybridization of phosphorylated O31; O32 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Hybridization of phosphorylated oligonucleotides: O31; O32 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2 µL O31 | ||
| + | * 2 µL O32 | ||
| + | * 200 µL ddH20 | ||
| + | * 37 °C 20 min | ||
| + | *75 °C 10 min | ||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, August 17'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Gelelectrophoresis of GSY7 === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of experiment: gelelectrophoresis of the remaining product of GSY7 PCR (from 16.8.16)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Agarose gelelectrophoresis was performed using an increased amount of SYBR Green | ||
| + | |||
| + | '''Results''' | ||
| + | There was no PCR product identified. | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Plasmidpreparation of liquid culture of P64; P73 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of P64; P73 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |P64 | ||
| + | |pSB1C3 | ||
| + | |VEGF-p2a-PDGF | ||
| + | |- | ||
| + | |P73 | ||
| + | |pSB1C3 | ||
| + | |Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * According to Mini-prep. kit, SLG | ||
| + | |||
| + | '''Results:'''' | ||
| + | * P64 134.4 ng/µL | ||
| + | * P73 149.4 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Over night liquid culture of F1015 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | ==='''PCR of scAvidin'''=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim:''' Extraction of single chain Avidin from vector for further cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * reaction was performed after SOP (PCR Using Q5® High-Fidelity DNA Polymerase, NEB) | ||
| + | * 2 ng Plasmid were used for amplification (1 µL) | ||
| + | * Primers: O-27 and O-28 | ||
| + | |||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial denaturation||98||120 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |64||30 | ||
| + | |- | ||
| + | |72||300 | ||
| + | |- | ||
| + | |final extension||72||300 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | === Quickchange of P206 and P239 to correct BAP=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim:''' Quickchange P206 (BAP-NanoLuc) and P239 (BAP-NanoLuc-A3C5) to correct BAP | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * reaction was performed after SOP (PCR usingPfuUltra II Fusion HS DNA Polymerase, Agilent) | ||
| + | * 10 ng Plasmid were used for amplification (2.5 µL) | ||
| + | * Primers: O-33 and O-34 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial denaturation||98||120 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |55||30 | ||
| + | |- | ||
| + | |68||300 | ||
| + | |- | ||
| + | |final extension||68||360 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inocculation of fermenter === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * centrifuge preculture (P258 in M9 medium) | ||
| + | * resuspend pellet in HCDF medium of fermenter and inocculate | ||
| + | * pO<sub>2</sub>= 60 %; Temp= 37°C, pH~7 | ||
| + | * growth over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, Aug 18th'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === PCR of gene synthesises 8 with Q5 polymerase === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of GSY7 with optimized Tm following negativ PCR reactions in order to amplify GSY7. | ||
| + | '''Procedure:''' | ||
| + | Approach: | ||
| + | |||
| + | * 2,5 µl forward Primer VF2 | ||
| + | *2,5µl reverse Primer VR | ||
| + | * 1µl dNTPs | ||
| + | * 1µl DNA (1ng) | ||
| + | * 10µl 5x Q5-Puffer | ||
| + | * 0,5µl Q5-Polymerase | ||
| + | * Fill up with water up to 50 µl | ||
| + | Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/) | ||
| + | * 98°C_2min initiation step | ||
| + | * 98°C_10sec | ||
| + | * 62°C_30sec > step 2-4: 35 repeats | ||
| + | * 72°C_1min | ||
| + | * 72°C_5min final step | ||
| + | * 4°_hold | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === PCR of gene synthesises 8 with Pfu Ultra II polymerase === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of GSY7 with Pfu Ultra II polymerase following negativ PCR reactions in order to amplify GSY7. | ||
| + | '''Procedure:''' | ||
| + | Approach: | ||
| + | |||
| + | * 2,5 µl forward Primer VF2 | ||
| + | *2,5µl reverse Primer VR | ||
| + | * 1µl dNTPs | ||
| + | * 1µl DNA (1ng) | ||
| + | * 10µl Pfu Ultra II-Puffer | ||
| + | * 0,5µl Q5-Polymerase | ||
| + | * Fill up with water up to 50 µl | ||
| + | Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/) | ||
| + | * 98°C_2min initiation step | ||
| + | * 98°C_10sec | ||
| + | * 56°C_30sec > step 2-4: 35 repeats | ||
| + | * 72°C_2min | ||
| + | * 72°C_5min final step | ||
| + | * 4°_hold | ||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Fermentation === | ||
| + | |||
| + | '''Investigator:''' NA | ||
| + | |||
| + | '''Aim of the experiment:''' Streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * exponential feed (Cam) for ~13 h (about 3-4 L); start at OD= 11.66 | ||
| + | * Induction of streptavidin expression (anhydro tetracyclin) at OD= 119 | ||
| + | * change of settings: pO2= 40 %; Temp= 30°C, pH~7 | ||
| + | * production over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Streptavidin production in shaking flask=== | ||
| + | |||
| + | '''Investigator:''' NA | ||
| + | |||
| + | '''Aim of the experiment:''' Streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2 L LB medium (Cam 1:1000) was inocculated with ~10 ml M9 preculture (P258 in BL21) | ||
| + | * induction with anhydro tetracycline (200µl, 2 mg/ml) at OD= 3,32 | ||
| + | * culture was incubated at 37 °C and 140 rpm for 4 hours | ||
| + | * after expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor) | ||
| + | * the supernatant was discarded, and the pellet was frozen away | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class= "receptor"> | ||
| + | |||
| + | ===Transformation of quickchanged P239 and 206 (BAP_Nanoluc_A3C5 and BAP_Nanoluc) === | ||
| + | |||
| + | '''Investigator: LK''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * After DpnI-digestion 10µl of each plasmid was transformed into E.coli XL-1-blue. | ||
| + | * Cells where plated out an Cam-plates after 1h of incubation without antibiotics. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class= "receptor"> | ||
| + | |||
| + | === MiniPrep of F209 to P266 and P267, analytic digestion === | ||
| + | |||
| + | '''Investigator: JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successful cloning. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) | ||
| + | |||
| + | * Concentrations: | ||
| + | <table cellspacing="0" border="1"> | ||
| + | <tr> | ||
| + | <td><b>Plasmid</b> | ||
| + | </td><td><b>c [ng/µl]</b> | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P266 | ||
| + | </td><td>198 | ||
| + | </td></tr> | ||
| + | <tr> | ||
| + | <td>P267 | ||
| + | </td><td>200,8 | ||
| + | </td></tr></table> | ||
| + | |||
| + | * Analytic digestion: 0.5 µl PstI, 0,5 µl EcoRI, 1 µl CutSmart, 6 µl ddH2O for P266 and P267 for 2 h at 37°C. | ||
| + | * 1% agarose gel electrophoresis. | ||
| + | |||
| + | '''Results:''' | ||
| + | * Lane 1: 10 µl 1 kb Ladder. | ||
| + | * Lane 2: Digest of F209 to P266. | ||
| + | * Lane 3: Digest of F209 to P267. | ||
| + | |||
| + | [[File:Muc16_miniPrep_of_P42_to_P50.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of O31; O32 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Phosphorylation of O31; O32 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL Oligonucleotides (µM) | ||
| + | * 2 µL 10x T4 PNK Bufer | ||
| + | * 2 µL 10 mM ATP | ||
| + | * 1 µL T4 PNK | ||
| + | * 10 µL H2O | ||
| + | * 95 °C 3min. | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Hybridization of phosphorylated O31; O32 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Hybridization of phosphorylated oligonucleotides: O31; O32 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2 µL O31 | ||
| + | * 2 µL O32 | ||
| + | * 200 µL ddH20 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Plasmidpreparation of liquid culture of F1015 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of F1015 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''#''' | ||
| + | |'''Plasmid''' | ||
| + | |'''Description''' | ||
| + | |- | ||
| + | |F1015 | ||
| + | |pSB1C3 | ||
| + | |VEGF-p2a-PDGF + Poly-A | ||
| + | |} | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * According to Mini-prep. kit, SLG | ||
| + | |||
| + | '''Results:'''' | ||
| + | * F1015 -> P1008 256.6 ng/µL | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, Aug 19th'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Gelelectrophoresis of GSY7 === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of experiment: gel electrophoresis of repeated GSY7 PCR (from 18.8.16)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Agarose gelelectrophoresis was performed at 90V | ||
| + | |||
| + | '''Results''' | ||
| + | There was no PCR product identified. | ||
| + | </div> | ||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === harvesting streptavidin === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * OD= 181 | ||
| + | * centrifuge culture for 1 h at 4800 rpm | ||
| + | * freeze pellet (2033 g) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === SDS PAGE Streptavidin fermentation === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verification of successfull production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * according to protocol | ||
| + | * 8 µl Ladder; before induction; after induction; sample of shaking flask production after induction | ||
| + | * all samples: 0.5/OD µl culture filled up with EB to 80µl + 20 µl SDS buffer; 10 µl on SDS gel | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16-deinemudder.jpg]] | ||
| + | |||
| + | |||
| + | |||
| + | Clear bands of acetyl transferase (chloramphenicol resistance) in fermenter samples. Therefore cells kept plasmid. The streptavidin bands before and after induction do not differ. The fermentation was not succesful. | ||
| + | The shaking flask sample however shows a brighter band for streptavidin. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Miniprep of F211 (pASK111(PAS Lys)), I739001 (Biobrick) and F206 (pASK111 vectorfragment) === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Miniprep of plasmids | ||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed by manufacturers protocol (Qiaprep, Qiagen) | ||
| + | * Plasmids were named: | ||
| + | ** F211 a: P268 | ||
| + | ** F211 b: P269 | ||
| + | ** I739001 a: P270 | ||
| + | ** I739001 b: P271 | ||
| + | ** F206 a: P272 | ||
| + | ** F206 b: P273 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Cloning of P260 in P261 and F215 in P74 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Digestion, ligation and transformation | ||
| + | '''Procedure:''' | ||
| + | |||
| + | * | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Transfection of HEK cells with P253, P225 and P226 === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Transfecting HEK cells with vectors encoding 3 diffrent types of signal peptides for secretion. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * HEK293 where cultured and seeded out on 12 cellculture dishes. (~5x10<sup>5</sup> cells per dish) | ||
| + | * Transfection was performed on day 3 after seeding (60-70% confluency) | ||
| + | * 10µg DNA of each signalpeptide mixed with 30µg PEI (polyenthylenimine) in ddH2O where incubated for 15 min and added to the cells, which where previously washed with PBS and covered (4mL) with optiMEM media. (Pen/Strep;No serum!) | ||
| + | * After 4h of incubation at 37°C, 5% CO<sub>2</sub> the media was changed to RPMI (Pen/Strep; 5% FCS) again. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Picking of ''E. coli'' XL1 blue in quickchanged pSB1C3 in P239 and P206, Retrafo of P261 and P213 === | ||
| + | |||
| + | '''Investigator: JL''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Quickchanged pSB1C3 in P239: Colonies were picked from Cam-plates. | ||
| + | |||
| + | * Retrafo of P261 and P213: Colonies were picked from Cam-plates. | ||
| + | |||
| + | * Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL of the proper antibiotic. | ||
| + | |||
| + | * These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | ==='''Repeat of PCR of scAvidin'''=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim:''' Extraction of single chain Avidin from vector for further cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * reaction was performed after SOP (PCR Using Q5® High-Fidelity 2X Master Mix, NEB) | ||
| + | * 2 ng Plasmid were used for amplification | ||
| + | * Primers: O-27 and O-28 | ||
| + | * annealing temperature was lowered to 60 °C | ||
| + | |||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial denaturation||98||120 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |60||30 | ||
| + | |- | ||
| + | |72||150 | ||
| + | |- | ||
| + | |final extension||72||300 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | ==='''Repeat of PCR of Gene Synthesis 9'''=== | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim:''' Amplification of gensynthesis 9 for further cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * reaction was performed after SOP (PCR Using Q5® High-Fidelity 2X Master Mix, NEB) | ||
| + | * 2 ng Plasmid were used for amplification | ||
| + | * Primers: VF2 and VR | ||
| + | |||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial denaturation||98||120 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |61||30 | ||
| + | |- | ||
| + | |72||120 | ||
| + | |- | ||
| + | |final extension||72||300 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Preparative digestion of P8 (Streptavidin wt) === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of experiment:''' preparation for cloning of SA1 into pASK111 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * preparative digestion batch | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |40 µl | ||
| + | |Plasmid DNA P8 | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Cutsmart (10x) | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |HindIII | ||
| + | |- | ||
| + | |1.5 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |2 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=50 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *duration: over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Saturday, Aug 20th'''== | ||
| + | |||
| + | <div class= "receptor"> | ||
| + | |||
| + | === Miniprep and analytical digestion of P274-P277 === | ||
| + | |||
| + | '''Investigator:''' LK | ||
| + | |||
| + | '''Aim of the experiment:''' Isolating and verifiing A3C5+EspP_ddT ligation, "quickchanged" BAP-constructs and amplification of A3C5 in pSB1C3. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Minipreps where performed according to manufacturer's protocol. | ||
| + | * Analytical digestion with EcoRI and PstI of ~ 200-250ng of each plasmid was performed for 30 min at 37°C. | ||
| + | |||
| + | '''Results:''' | ||
| + | * | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Sunday, Aug 21st'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Cloning of GSY9 and scAvidin === | ||
| + | |||
| + | '''Investigaor:''' LK, JL | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * The PCR of scAvdin was digested over night with SpeI and NgoMIV at 37°C. | ||
| + | * The PCR of GSY9 was divided: one half was digested with XbaI and AgeI (-> F224) and the other half was digested with XbaI and HindIII (->F225). | ||
| + | * Additionally P74 (pSB1C3+RFP-Reporter) was digested with XbaI and AgeI (-> F227) to provide a plasmid for the ligation with F224. | ||
| + | |||
| + | * The digestions where plotted on a gel, and bands at 1700Bp for scAvidin, 222Bp for TetO-OmpA, 490Bp for eMA and 2100Bp for pSB1C3 where cut out and purified with the Promega Wizard SV Gel Kit according to the manufactuerers protocol. | ||
| + | |||
| + | * F224 (TetO-OmpA) was then ligated with F227 (pSB1C3), F225 (enhanced monomeric Avidin) was ligated with F206 (pASK111) and F226 (scAvidin) was ligated with F218 (pSB1C3). Ligations where incubated over night at 16°C. | ||
| + | |||
| + | * Transformation was performed according to the SOP on Monday morning. | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | =Week 15, Aug 22nd - Aug 28th= | ||
| + | |||
| + | <div class="week" id="Week_15"> | ||
| + | |||
| + | =='''Monday, Aug 22nd'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Sequencing of P268, P276, P277 and P279 === | ||
| + | |||
| + | '''Investigator: Julian''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P268 (PAS lysine), P276 (QC BAP-NanoLuc-A3C5), P277 (QC BAP-NanoLuc) and P279 (A3C5-EspP-DoubleTerminator) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) | ||
| + | * P268 (D20) FR12904429 | ||
| + | * P276 (VF2) FR12904428 | ||
| + | * P277 (VF2) FR12904427 | ||
| + | * P279 (VF2) FR12904426 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion of P278 (pSB1C3(A3C5-EspP-doubleterminator) & P279 === | ||
| + | |||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | |||
| + | '''Aim of the experiment:''' cofirm success of cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Batch for analytical digestion for P278/P279 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl/ 1 µl | ||
| + | |Plasmid DNA P278/P279 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |5 µl/ 6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * P279 showed the expected signal at 1100 bp | ||
| + | |||
| + | </div> | ||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligations of F1013 + gsy7; +gsy8 === | ||
| + | |||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | |||
| + | '''Aim of the experiment:''' cloning for further ligation steps | ||
| + | '''Procedure:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, Aug 23rd'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Inoculation of Precultures of F228, F229 and F230 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Prepare cells for Miniprep | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Two colonies were picked from each plate | ||
| + | * LB CAM was inoculated with F228 and F230 | ||
| + | * LB AMP was inoculated with F229 | ||
| + | * Precultures were incubated for 7 hours at 37°C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Each preculture showed cell growth | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Miniprep of F228, F229 and F30 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Obtain plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN) | ||
| + | |||
| + | * Plasmids were renamed: | ||
| + | ** F228 1: P281 | ||
| + | ** F228 2: P282 | ||
| + | ** F229 1: P283 | ||
| + | ** F229 2: P284 | ||
| + | ** F230 1: P285 | ||
| + | ** F230 2: P286 | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Concetrations in "Inventarliste" | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion of IRES-construct (F1019) by PmeI+NotI === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Prepare construct for ligation later on | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 5 µl DNA | ||
| + | 38µl ddH2O | ||
| + | 1µl of each enzyme | ||
| + | 5µl cut smart buffer | ||
| + | |||
| + | incubation at 37°C for 1 h | ||
| + | Gel electrophoresis and gel extraction | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Fragment was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, Aug 24th'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === analytical digestion of P281, P282, P283, P284, P285 and P286 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Verify success of cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | * Batch for analytical digestion of P281/P282 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |Plasmid DNA P281/P282 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |6 µl/ 6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P283/P284 with XbaI and HindIII-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |Plasmid DNA P283/P284 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |HindIII-HF | ||
| + | |- | ||
| + | |6 µl/ 6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P285/P286 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 1 µl | ||
| + | |Plasmid DNA P285/P286 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |6 µl/ 6 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated at 37° for 1h | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === PCR of Genesynthesis 9 with VF2 and VR === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' Amplification of GSY9 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * reaction was performed after SOP (Q5 Hotstart, NEB) | ||
| + | * 2 ng Plasmid were used for amplification | ||
| + | * Primers: VF2 and VR | ||
| + | * Over all volume= 50 µl | ||
| + | |||
| + | *PCR program (standard): | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Step''' | ||
| + | |'''Temperature [°C]''' | ||
| + | |'''Time [s]''' | ||
| + | |- | ||
| + | |initial denaturation||98||120 | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98||10 | ||
| + | |- | ||
| + | |66||30 | ||
| + | |- | ||
| + | |72||30 | ||
| + | |- | ||
| + | |final extension||72||300 | ||
| + | |- | ||
| + | |Hold||4||ꝏ | ||
| + | |} | ||
| + | |||
| + | * PCR purification was performed according to manufacturers protocol (Wizard SV Gel and PCR clean-up kit, Promega) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === PCR of linear pSB1C3 === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of linear pSB1C3 and introduction of restriction site: blunt at 5' and NotI at 3' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (O-038) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (O-039) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |1 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |68 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion of P74, P165, P196, P273, P274, P275, P285 and PCR of GSY9 === | ||
| + | |||
| + | '''Investigator: Clemens''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | * Batch for preparative digestion of P74 with XbaI and AgeI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Plasmid DNA P74 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |23 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P74 with NgOMIV and AgeI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2 µl | ||
| + | |Plasmid DNA P74 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |23 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P165 with PstI-HF and AgeI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |12 µl | ||
| + | |Plasmid DNA P165 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |13 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P196 with NgOMIV and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA P196 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |10 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P273 with XbaI and HindIII-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |20 µl | ||
| + | |Plasmid DNA P273 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |HindIII-HF | ||
| + | |- | ||
| + | |5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P274 with AgeI-HF and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA P274 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P275 with NgOMIV and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |8 µl | ||
| + | |Plasmid DNA P275 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |17 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P285 with NgOMIV and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |12 µl | ||
| + | |Plasmid DNA P285 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |13 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of PCR of GS9 with NgOMIV, AgeI-HF and XbaI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |25 µl | ||
| + | |Plasmid DNA PCR GSY9 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |0 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=31 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of PCR of GS9 with HindIII-HF and XbaI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |25 µl | ||
| + | |Plasmid DNA PCR GSY9 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |HindIII-HF | ||
| + | |- | ||
| + | |0 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated at 37° over night | ||
| + | * Samples were applied on a 1% agarose gel | ||
| + | * Samples were seperated for 45 min at 80V | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * All Digests showed the expected lengths | ||
| + | * Fragments were cut from the gel and purified according to manufacturers protocol (Wizard SV Gel and PCR clean-up system, Promega) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Transformation of P296 (pSB1C3 mGMK-Tk30, BBa_K782063) in xl1-blue === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of the experiment:''' Transformation for Poly-A-tail adding | ||
| + | |||
| + | '''Procedure:''' | ||
| + | transformation in competent xl1-blue according to protocol + rescue | ||
| + | |||
| + | '''Result:''' | ||
| + | plates (LB Cam) in incubator for further processing (37 °C) showed after 24 h colonies | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === (Re)ligations of F1013 (pSB1C3) and F1016 (phosphoryliert) + F1013 and F1005 in pSB1C3 === | ||
| + | |||
| + | '''Investigator: EF''' | ||
| + | |||
| + | '''Aim of the experiment:''' Ligations of PCR Produkt from GSY8 in pSB1C3 vector for later transformation and cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Ligation mixtures were prepared as following; DNA amounts were calculated using the ligation calculator http://www.insilico.uni-duesseldorf.de/Lig_Input.html : | ||
| + | * F1013+F1016: 1.6 μL F1013, 5 μL F1016, 2μL T4-DNA-Ligase-Buffer, 1 μL T4-DNA-Ligase, 10.4 μL ddH2O | ||
| + | * F1013+F1005: 1.6 μL F1013, 2.2 μL F1005, 2μL T4-DNA-Ligase-Buffer, 1 μL T4-DNA-Ligase, 13.2 ddH2O | ||
| + | * Ligation was incubated for 1.5 hours at 16 °C | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="Hypoxia"> | ||
| + | |||
| + | =='''Thursday, Aug 25th'''== | ||
| + | |||
| + | </div> | ||
| + | <div class="general"> | ||
| + | |||
| + | === Phosphorylation and hybridization of O35, O36, O37 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Phosphorylation and hybridization of O35, O36, O37 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | Phosphorylation | ||
| + | * 0,5 µL Oligonucleotides | ||
| + | * 2 µL 10x T4 PNK Bufer | ||
| + | * 2 µL T4 Ligase Buffer | ||
| + | * 1 µL T4 PNK | ||
| + | * 14,5 µL H2O | ||
| + | incubate at 37°C for 20 min and inactivate T4PNK (for 10 min at 75°C) | ||
| + | |||
| + | |||
| + | Hybridization | ||
| + | * 2 µL O31 | ||
| + | * 2 µL O32 | ||
| + | * in 200 µL ddH20 | ||
| + | incubate at 95°C for 3 min, let cool down slowly | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Inoculation of P296(Kill-Switch) === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' production of P296 | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | 2x5ml LB-CAM media, each inoculated by one colony (not by rescue colonies) | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P74 + HincII === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P74 + HincII | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA (1431 ng/µL) | ||
| + | * 1 µL HincII | ||
| + | * 14 µL ddH2O | ||
| + | * 2 µL CutSmart buffer | ||
| + | * incubation for 2h at 37 °C | ||
| + | |||
| + | '''Results:'''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_P74_HincII.png|250px]] | ||
| + | |||
| + | * P74 -> P1021 111,2 ng/µL | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligation of F233+F238, F236+F234, F233+F237, F232+F240, F231+F239, F235+F241 and F248+F249 === | ||
| + | |||
| + | '''Investigator: ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Ligation and transformation of XL1 blue | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Batch for Ligation of F233 and F238 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1,7 µl | ||
| + | |Vector F233 | ||
| + | |- | ||
| + | |8,3 µl | ||
| + | |Insert F238 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F236 and F234 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1,1 µl | ||
| + | |Vector F236 | ||
| + | |- | ||
| + | |8,9 µl | ||
| + | |Insert F234 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F233 and F237 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3,1 µl | ||
| + | |Vector F233 | ||
| + | |- | ||
| + | |6,7 µl | ||
| + | |Insert F237 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F232 and F240 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |4,8 µl | ||
| + | |Vector F232 | ||
| + | |- | ||
| + | |5,2 µl | ||
| + | |Insert F240 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F231 and F239 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |5,6 µl | ||
| + | |Vector F231 | ||
| + | |- | ||
| + | |4,4 µl | ||
| + | |Insert F239 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F235 and F241 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6,5 µl | ||
| + | |Vector F235 | ||
| + | |- | ||
| + | |3,5 µl | ||
| + | |Insert F241 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F228 and F249 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2,7 µl | ||
| + | |Vector F248 | ||
| + | |- | ||
| + | |7,3 µl | ||
| + | |Insert F249 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Ligations were performed according to SOP | ||
| + | * XL1blue were transformed with the resulting fragments | ||
| + | |||
| + | |||
| + | '''Results:'''' | ||
| + | |||
| + | Cellgrowth yes/no? | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P1007 + EcoRI + SpeI=== | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P1007 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 10 µL DNA (3.6 ug) | ||
| + | * 0.25 µL EcoRI | ||
| + | * 0.25 µL SpeI | ||
| + | * 34.5 µL ddH2O | ||
| + | * 5 µL CutSmart Buffer NEB | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion, gel electrophoresis and gel extraction of P288 (CMV-Promotor) and P289 (NanoLuc + Streptag + Poly-A) === | ||
| + | |||
| + | '''Investigator: JL''' | ||
| + | |||
| + | '''Aim of experiment: Preparative digestion, gel electrophoresis and gel extraction of P288 (CMV-Promotor) and P289 (NanoLuc + Streptag + Poly-A)''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batches for analytical digestions: | ||
| + | |||
| + | *P288: PstI and SpeI. | ||
| + | |||
| + | *P29: PstI and XbaI. | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |3 µl | ||
| + | |CutSmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Each enzyme | ||
| + | |- | ||
| + | |30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | *Preparative gel electrophoresis was performed at 90 V for 60 min. | ||
| + | *Preperative gel according to SOP. | ||
| + | *The Purification was done according to the manufacturer's protocol. | ||
| + | |||
| + | [[File:MUC16_gelextraction_P288_P289.jpeg]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of P293, P294, P295 (eMA pASK111) === | ||
| + | |||
| + | '''Investigator: JL''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analytical digestion and gelelectrophoresis of P293, P294, P295 (eMA pASK111). | ||
| + | |||
| + | |||
| + | '''Procedure:''' | ||
| + | * Batch for analytical digestion for P293, P294, P295. | ||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |2/1/4 µl | ||
| + | |Plasmid DNA P293/P294/P295 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |NEBuffer 4 (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |HindIII (20 U/µl) | ||
| + | |- | ||
| + | |6/8/4 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=12 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Incubation for 120 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 30 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |1 kbp ladder DNA ladder | ||
| + | |'''P293''' | ||
| + | |'''P294''' | ||
| + | |'''P294''' | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[File:TUM13_20130423_RFP_Generator_RFC25_AgeI_NgoMIV.png|500px]] | ||
| + | |||
| + | </div> | ||
| + | =='''Friday, Aug 26'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P74 + HincII === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P74 + HincII | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA (1431 ng/µL) | ||
| + | * 1 µL HincII | ||
| + | * 41 µL ddH2O | ||
| + | * 5 µL CutSmart buffer | ||
| + | * * 2h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_gelextraction_P74_HincII_2.png|250px]] | ||
| + | |||
| + | generuler; pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2; P74[HincII] | ||
| + | |||
| + | * F1024 49.3 ng\µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of Vector | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 + PmeI & NotI | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 7 µL DNA (77 ng/µL) | ||
| + | * 0.25 µL PmeI | ||
| + | * 0.25 µL NotI | ||
| + | * 34.5 µL ddH2O | ||
| + | * 5 µL CutSmart buffer | ||
| + | * 2h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_gelextraction_P74_HincII_2.png|250px]] | ||
| + | |||
| + | generuler; pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2; P74[HincII] | ||
| + | |||
| + | * F1023 7.6 ng\µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Gelextraction PCR pSB1C3 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Purification of PCR linear pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * According to Gelextraction kit; Quiagen | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_gelextraction_PCR_linear_pSB1C3.png|250px]] | ||
| + | |||
| + | generuler; PCR gsy8; PCR linear pSB1C3 | ||
| + | |||
| + | * F1025 9.2 ng\µL | ||
| + | </div> | ||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Analytical digestion of P249, P268 and P269 (each by XbaI&HindIII) === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analyze construct for eventual preparative digestion | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 5 µl DNA (except for P249 --> 13 µl) | ||
| + | 12µl ddH2O (except for P249 --> 4 µl) | ||
| + | 1µl of each enzyme | ||
| + | 2µl cut smart buffer | ||
| + | |||
| + | incubation at 37°C for 2 h | ||
| + | Gel electrophoresis and gel extraction | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | In each digestion only one fragment (the vector) was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder. | ||
| + | |||
| + | </div> | ||
| + | <div class="linkers"> | ||
| + | |||
| + | === Preparative digestion of P272 and P273 (each by XbaI&HindIII) === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Prepare construct for ligation later on | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 5 µl DNA | ||
| + | 36µl ddH2O | ||
| + | 2µl of each enzyme | ||
| + | 5µl cut smart buffer | ||
| + | |||
| + | incubation at 37°C for 2 h | ||
| + | Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Fragment was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Miniprep of P296 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Obtain plasmids | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN) | ||
| + | |||
| + | * Plasmid was not renamed. | ||
| + | ** | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Concetrations in "Inventarliste" | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | === Sequencing of P296 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P296 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Sequencing batch was prepared after manufacturer's protocol: | ||
| + | |||
| + | **7 µl of plasmid DNA (end concentration 50 - 100ng) in 8µl H2O | ||
| + | **2 µl sequencing primer (one batch with VF2 and another with VR) | ||
| + | |||
| + | *The different vectors we sequenced received the following barcodes: | ||
| + | |||
| + | ** P296 with VF2: FR | ||
| + | ** P296 with VR: FR | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion of P1007, P1008 with EcoRI HF, SpeI HF + analytical gel === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Isolation of pSB1C3 plasmid from p1007) | ||
| + | '''Procedure:''' | ||
| + | |||
| + | * p1007 digested with: | ||
| + | 0,25 µl EcoRI (HF) | ||
| + | 0,25 µl SpeI (HF), | ||
| + | 5 µl CutSmart Buffer | ||
| + | 3 µg plasmid-DNA (P1007) | ||
| + | fill up with ddH2O (Vtotal= 50µL) | ||
| + | |||
| + | * 2x 25 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard) | ||
| + | |||
| + | '''Results:''' Despite optimised enzyme quantity the insert seems to not have been cut out of the plasmid. One single band at 3kb --> repeat reaction; check cutting sites with genious | ||
| + | (*)has p1007 been sequenzed before(?). However the band was extracted from the geled and freezed labeled as "8.1" | ||
| + | |||
| + | * p1008 digested with: | ||
| + | 0,25 µl EcoRI (HF) | ||
| + | 0,25 µl SpeI (HF), | ||
| + | 5 µl CutSmart Buffer | ||
| + | 3 µg plasmid-DNA (P1008) | ||
| + | fill up with ddH2O (Vtotal= 50µL) | ||
| + | |||
| + | * 2x 25 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard) | ||
| + | |||
| + | '''Results:''' Insert was isolated by gel extraction following GelExtraction Kit instructions. | ||
| + | |||
| + | |||
| + | [[File:P1007, P1008_26.8.16_EF_black.scn|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Saturday, Aug 27'''== | ||
| + | <div class="hypoxia"> | ||
| + | === PCR GSY8 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (VR) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (D20) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |61 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * negativ two times :( | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Mung Bean nuclease (MBN) digestion of F1023 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Get rid of overhangs. Preparation of DNA for blunt ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 18 µL DNA (144 ng) | ||
| + | * 2 µL MBN buffer | ||
| + | * 0.5 µL MBN | ||
| + | * 30 min; 30 °C | ||
| + | * purification by PCR purification kit, Promega | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1026 24 ng\µL; must be wrong, because this is more than digested. Endvolume 20 µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Dephosphorylation of F1024 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Preparation of Vekor for blunt end Ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 13 µL DNA (~50 ng\µL) | ||
| + | * 5 µL Antarctic phosphatase (AP) | ||
| + | * 2 µL AP buffer | ||
| + | * 30 min ; 37 °C | ||
| + | * heat inactivation: 2 min; 80 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1029 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation: F1025 +F1022 = F1027 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F1025 +F1022 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1 µL F1025 (10 ng\µL) | ||
| + | * 10 µL F1022 (5 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 6 µL ddH2O | ||
| + | * 60 min; 30°C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1025 +F1022 = F1027 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation: F1025 +F1026 = F1028 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F1025 +F1026 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1 µL F1025 (10 ng\µL) | ||
| + | * 20 µL F1026 (24 ng\µL) | ||
| + | * 3 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 4 µL ddH2O | ||
| + | * 60 min; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1025 +F1026 = F1028 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of GSY7; GSY8 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Phosphorylation of GSY7; GSY8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL DNA | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 1 µL T4 PNK | ||
| + | * 12 µL ddH20 | ||
| + | * 30 min; 37 °C | ||
| + | * heat inactivation: 20 min; 65 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1030 (GSY7) | ||
| + | * F1031 (GSY8) | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Over night liquid culture of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation: F1029 + F1030 = F1032; F1029 + F1031 = F1033 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Ligation: F1029 + F1030 = F1032; F1029 + F1031 = F1033 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL F1029 ( ~ 30 ng) {ERROR!} | ||
| + | * 10 µL F1030/F1031 (~ 25 ng) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 2 µL ddH2O | ||
| + | * 60 min; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1029 + F1030 = F1032 | ||
| + | * F1029 + F1031 = F1033 | ||
| + | *Both negativ, due to miscalculation | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F1027; F1028; F1032; F1033 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of Vector | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | =='''Sunday, Aug 28'''== | ||
| + | <div class="hypoxia"> | ||
| + | === PCR GSY8 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (D20) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (VR) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |58 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | '''Results:''' | ||
| + | * | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Over night liquid culture of F1032; F1028 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Plasmidpreparation of liquid culture of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 === | ||
| + | |||
| + | '''Investigator: MP ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * According to Mini-prep. kit, SLG | ||
| + | |||
| + | '''Results:'''' | ||
| + | * P1009 608.6 ng/µL | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | =Week 16, Aug 29nd - Sep 4th= | ||
| + | <div class="week" id="Week_16"> | ||
| + | |||
| + | =='''Monday, Aug 29nd'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Plasmid preparation of liquid culture of F1032(clon 1); F1032(clon 2) === | ||
| + | |||
| + | '''Investigator: JL ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Purification of F1032(clon 1); F1032(clon 2) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * According to Mini-prep. kit, Quiagen | ||
| + | |||
| + | '''Results:''' | ||
| + | * P 1010 448.4 ng/µL | ||
| + | * P 1011 344.6 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Analytical digestion of P1010; P1011 === | ||
| + | |||
| + | '''Investigator: JL ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * verify constructs: P1010; P1011 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2 µL DNA | ||
| + | * 0.5 µL EcoRI | ||
| + | * 0.5 µL PstI | ||
| + | * 1 µL CutSmart buffer | ||
| + | * 6 µL ddH20 | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_analytical_digest_P1010_P1011.png|250px]] | ||
| + | |||
| + | * 1kb ladder; P1010; P1011 | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === PCR and purification of GSY7_new and GSY8 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 7_new and GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | Same procedure for both GSYs: | ||
| + | |||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (VR) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (D20) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |61 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Purification according to Promega PCR purification kit | ||
| + | |||
| + | '''Results:''' | ||
| + | GSY7: 9,4 ng/µl | ||
| + | GSY8: 1,5 ng/µl, probably contaminated with GSY7_new | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Phosphorylation of GSY7_new and GSY8 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Phosphorylation of GSY7_new; GSY8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL DNA | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 1 µL T4 PNK | ||
| + | * 12 µL ddH20 | ||
| + | * 30 min; 37 °C | ||
| + | * heat inactivation: 10 min; 75 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1034 (GSY7_new) | ||
| + | * F1035 (GSY8) | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F1034 + F1029 = F1036; F1035 + F1029 = F1037 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Ligation: F1034 + F1029 = F1036; F1035 + F1029 = F1037 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL F1029 ( ~ 10 ng) | ||
| + | * 10 µL F1034/F1035 (~ 25 ng) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 2 µL ddH2O | ||
| + | * 60 min; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1029 + F1034 = F1036 | ||
| + | * F1029 + F1035 = F1037 | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion of P297, P299, P279, P285, P285, P277, P312 & P251 === | ||
| + | |||
| + | '''Investigator: ''' Clemens | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Preparation for cloning in pSB1C3/pASK111 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * * Batch for preparative digestion of P297 with AgeI-HF & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Plasmid DNA P297 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |220 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P299 with NgOMIV and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P299 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P279 with NgOMIV & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |20 µl | ||
| + | |Plasmid DNA P279 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * * Batch for preparative digestion of P285 with AgeI-HF & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA P297 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |10 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * * Batch for preparative digestion of P277 with AgeI-HF & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P277 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * * Batch for preparative digestion of P312 with AgeI-HF & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |5 µl | ||
| + | |Plasmid DNA P312 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |20 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P251 with NgOMIV & PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA P251 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |10 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated for 3h at 37°C | ||
| + | * Fragments were cut from the gel and purified by gelectraction (promega) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Fragments were renamed (see Inventarliste) and concentrations were measured | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligation of F254+ F255, F256+F257, F256+F258 & F260+F312 === | ||
| + | |||
| + | '''Investigator: ''' Clemens | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Batch for Ligation of F254 and F255 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Vector F254 | ||
| + | |- | ||
| + | |7 µl | ||
| + | |Insert F255 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F257 and F256 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.9 µl | ||
| + | |Vector F257 | ||
| + | |- | ||
| + | |8.1 µl | ||
| + | |Insert F256 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F258 and F256 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.3 µl | ||
| + | |Vector F258 | ||
| + | |- | ||
| + | |8.7 µl | ||
| + | |Insert F256 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | *Batch for Ligation of F312 and F260 | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1.2 µl | ||
| + | |Vector F312 | ||
| + | |- | ||
| + | |8.8 µl | ||
| + | |Insert F260 | ||
| + | |- | ||
| + | |2 µl | ||
| + | |10X DNA-Ligase-buffer | ||
| + | |- | ||
| + | |1 µl | ||
| + | |T4-Ligase | ||
| + | |- | ||
| + | |7 µl | ||
| + | |H₂O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | * Ligations were performed according to SOP | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Analytical digestion and gelelectrophoresis of pASK111, P268 (SUMO-PAS), P6, and P259 (both SA) === | ||
| + | |||
| + | '''Investigator: Niklas''' | ||
| + | |||
| + | '''Aim of the experiment: verification before cloning for fermentation''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''P268/ P259/ P6/ pASK111''' | ||
| + | |- | ||
| + | |5,8 µl/ 17 µl/ 8,33 µl/ 14,2 µl | ||
| + | |Plasmid DNA | ||
| + | |- | ||
| + | |2 µl | ||
| + | |cut smart (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |HindIII | ||
| + | |- | ||
| + | |11,2 µl/ 0 µl/ 8,67 µl/ 2,8 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=20 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | * Incubation for 90 min at 37 °C. | ||
| + | |||
| + | * Analytical gelelectrophoresis was performed at 90 V for 60 min. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_asdfghjklmnbv.jpg]] | ||
| + | |||
| + | * on of the enzymes was contaminated | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Transformation of P310 in BL 21; P6, P259, P268, pASK111, F1034+F1028 in xl1-blue === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * CaCl<sub>2</sub>-competent ''E. coli'' cells were taken from the stock in –80 °C freezer and gently thawed on ice | ||
| + | |||
| + | * 1 µl (retrafo)/ 7 µl (ligation) of DNA was added to 50 µl of competent cells and gently mixed | ||
| + | |||
| + | * 30 min incubation on ice | ||
| + | |||
| + | * 5 min heat shock at 37 °C | ||
| + | |||
| + | * Added 0,9 ml LB medium to each tube | ||
| + | |||
| + | * Incubation for 60 min at 37 °C in the 180 rpm cell-culture shaker | ||
| + | |||
| + | * 50 µl of the cell suspension was plated on one plate | ||
| + | |||
| + | * The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded | ||
| + | |||
| + | * The pellet was resuspended in 50 µl of LB-medium and this concentrated cell suspension was plated on a plate. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, Aug 30rd'''== | ||
| + | <div class="general"> | ||
| + | === PCR on GSY7 and GSY8 === | ||
| + | |||
| + | '''Investigator: CG ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 7 and GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | Same procedure for both GSYs: | ||
| + | |||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (VR) | ||
| + | |1,5 μl | ||
| + | |0.25 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (D20) | ||
| + | |1,5 μl | ||
| + | |0.25 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |2 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|45 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |60 °C | ||
| + | |90 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | * GSY8 negative: smear at the upper end of the gel. | ||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion of P270 & P308 === | ||
| + | |||
| + | '''Investigator: ''' Clemens | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Preparation for cloning in pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Batch for preparative digestion of P270 with EcoRI-HF & Spe-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |25 µl | ||
| + | |Plasmid DNA P270 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |SpeI-HF | ||
| + | |- | ||
| + | |0 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P308 with EcoRI-HF and XbaI | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |10 µl | ||
| + | |Plasmid DNA P308 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |15 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | |||
| + | * Digestions were incubated for 3h at 37°C | ||
| + | * Fragments were cut from the gel and purified by gelectraction (promega) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Fragments were renamed (see Inventarliste) and concentrations were measured | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Cloning of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) in pSB1C3 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Cloning of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) in pSB1C3 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 17 µl pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) digestion with 1 µl Mung Bean Nuclease and 2 µl buffer for 30 min. at 30°C to receive blunt ends | ||
| + | * Ligation of 8.7 µl pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) and 1.3 µl pSB1C3 | ||
| + | * Overnight incubation at 16 °C | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Retransformation of P258 === | ||
| + | |||
| + | '''Investigator: CG ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of Vector | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Digestion of P1005, P1006, P1007 with XbaI, PstI HF + analytical gel === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Isolation of insert for further construction of hypoxia system | ||
| + | '''Procedure:''' | ||
| + | |||
| + | * p1005, p1006, p1007 digested with: | ||
| + | 0,25 µl EcoRI (HF) | ||
| + | 0,25 µl SpeI (HF), | ||
| + | 5 µl CutSmart Buffer | ||
| + | 3 µg plasmid-DNA (Pp1005, p1006, p1007) | ||
| + | fill up with ddH2O (Vtotal= 50µL) | ||
| + | |||
| + | * 30 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard) | ||
| + | |||
| + | * Digestions were incubated for 1h at 37°C | ||
| + | |||
| + | '''Results:''' Insert and vector were clearly separated. All cut vector remained at the expected position of 2kb. Inserts were isolated following extraction kit protocol . F number and concentrations were included into the Inventarliste. Remain of 30µl (with dye) frozen at -20 C -> Labeled as 5.1; 6,1; 7.2 | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Digestion of P1007 with EcoRI HF, SpeI HF + analytical gel === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' Isolation of vector. Verification of digestion performed on 29th, Friday | ||
| + | '''Procedure:''' | ||
| + | |||
| + | *p1007 digested with: | ||
| + | 0,25 µl EcoRI (HF) | ||
| + | 0,25 µl SpeI (HF), | ||
| + | 5 µl CutSmart Buffer | ||
| + | 3 µg plasmid-DNA (p1007) | ||
| + | fill up with ddH2O (Vtotal= 50µL) | ||
| + | |||
| + | * Digestion was incubated for 1h at 37°C | ||
| + | |||
| + | * 30 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard) | ||
| + | |||
| + | '''Results:''' Again plasmid wasn´t cut. One single band at 3kb. Further procedur: sequence plasmid (?) repeat ligation to generate p1007. Isolate pSB1C3 from another plasmid. | ||
| + | |||
| + | |||
| + | |||
| + | [File to be uploaded] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Ligation of F1001 and F1002 === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' generate p1007 due to gel results (since 28th, Thursday) | ||
| + | '''Procedure:''' Ligation was performed as follows: 6,3 µl F1001, 5,3 µl F1002, 2 µl T4-DNA-Ligase Buffer, 1 µl T4-DNA-Ligase, filled up with ddH20 to 20 µl *3:1 vector-insert ratio | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation of F248 and 1015 === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | '''Aim of the experiment:'' hypoxia | ||
| + | '''Procedure:''' Ligation was performed as follows: 1 µl F248, 5 µl F1015, 2 µl T4-DNA-Ligase Buffer, 1 µl T4-DNA-Ligase, filled up with ddH20 to 20 µl *Approximate 3:1 vector insert ratio | ||
| + | |||
| + | '''Results''': Plates incubating over night at 37C | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Sequencing of P259 (SA from GSY6; P303 and P304 (eMA) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) - all samples: VF2 | ||
| + | * barcodes in stock list | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Picking of of ''E. coli'' XL1 blue with P6, pASK111, P268 === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 2 colonies were picked. | ||
| + | |||
| + | * These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inocculation of preculture (P310 eMA-HisTag) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 2 colonies were picked (20 mL LB with Cam) | ||
| + | |||
| + | * the preculture was transferred in a cell culture shaker at 37 °C and were incubated overnight | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting SUMO Protease=== | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' SUMO Protease production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 6 x 2 L cultures | ||
| + | * centrifuge cultures for 30 min at 6000 rpm | ||
| + | * freeze pellet (120.5 g) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, Aug 31st'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Over night liquid cultures of P1007 and P1012 === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Obtaining enough cellmass for plasmid preparation. Liquid cultures of Turbo and XL1 blue cells. | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of F1036 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Phosphorylation of F1036 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 36 µL DNA (*350 ng) | ||
| + | * 4 µL T4 ligase buffer | ||
| + | * 1 µL PNK | ||
| + | * 30 min; 37 °C | ||
| + | * heat deactivation: 20 min; 65 °C | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F1029 + F1037 = F1038 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Ligation: F1029 + F1037 = F1038 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 0.5 µL F1029 (30 ng\µL) | ||
| + | * 10 µL F1037 (9 ng\µL) | ||
| + | * 2 µL T4 ligase buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 7 µL ddH2O | ||
| + | * over night ; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1029 + F1037 = F1038 | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | === Miniprep of various plasmids, analytical digestion, sequencing === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Obtain plasmids for: eMA, PAS-Lys-Sumo, pASK111, Streptavidin (non-codon-optimized) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN) | ||
| + | * Digestion of about 250 ng for each eMA plasmid with EcoRI and PstI | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Names and concetrations in "Inventarliste" | ||
| + | * Sequencing results | ||
| + | * Gel: | ||
| + | 1. GSY8, 2. - 5.: P319-P322 with EcoRI / PstI | ||
| + | |||
| + | |||
| + | [[File:Muc16_160831_CG1.jpeg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Cloning of Streptavidin (non-codon-optimized) into pASK111 === | ||
| + | |||
| + | '''Investigator: JL, NA, CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Obtain Streptavidin (non-codon-optimized) in expression vector | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Preparative digestion of 42 µl P6 (Streptavidin) and 42 µl P315, 5 µl CutSmart and 1.5 µl XbaI/HindIII | ||
| + | * Gel extraction | ||
| + | * Ligation: 5.9 µl pASK111 + 4.1 µl SA + 2 µl ligase buffer + 7 µl ddH2o + 1 µl T4 ligase | ||
| + | * Transformation according to SOP | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Gel: | ||
| + | 1. Streptavidin (non-codon-optimized), 2. pASK111 | ||
| + | |||
| + | [[File:Muc16_160831_CG2.jpeg|500px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Preparative digestion of P296 and P73 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Prepare construct for ligation later on | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 5 µl DNA | ||
| + | 36µl ddH2O | ||
| + | 2µl of each enzyme (P296 --> EcoRI&SpeI, P73 --> EcoRI&XbaI) | ||
| + | 5µl cut smart buffer | ||
| + | |||
| + | incubation at 37°C for 3 h | ||
| + | Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Fragments were expected at about 1 kb (P296) and 2,5 kb (P73). This could be shown comparing with 1kb DNA ladder. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | === Ligation: F269 + F270 = F271 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Ligation: F269 + F270 = F271 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL F269 ( ~ ng) | ||
| + | * 10 µL F270 (~ ng) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 2 µL ddH2O | ||
| + | * ONT; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F269 + F270 = F271 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === PCR and purification of GSY7_new and GSY8 === | ||
| + | |||
| + | '''Investigator: CG, EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 7_new and GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | Same procedure for both GSYs: | ||
| + | |||
| + | * PCR using PfuUltra II Fusion HotStart DNA Polymerase according to manufacturer’s online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''10 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |PfuUltra II Hotstart PCR Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM Forward Primer (O-038) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (O-039) | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |2 min | ||
| + | |- | ||
| + | |rowspan = "3"|40 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |98 °C | ||
| + | |20 sec | ||
| + | |- | ||
| + | |45-64,7 °C (gradient) | ||
| + | |20 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Purification according to Promega PCR purification kit | ||
| + | |||
| + | '''Results:''' | ||
| + | no amplification recognizable | ||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting SUMO Protease=== | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' SUMO Protease production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * resuspend pellet in PBS + 10 mM EDTA | ||
| + | * homogenize cells with PANDA | ||
| + | * centrifuge cultures for 30 min at 8000 rpm (protein in supernantant) | ||
| + | * dialysis against IMAC buffer | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Inocculation and induction of eMA-HisTag (P310) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 2 L LB shaking flask | ||
| + | |||
| + | * the cell culture was incubated at 37 °C | ||
| + | * induction with 200 µl anhydro tetracycliine (2mg/ml) at OD= 0,55 | ||
| + | * production over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, Sep 1st'''== | ||
| + | <div class="receptor"> | ||
| + | === Miniprep of F261, F262, F263, F264, I739001, P305 & P258 ,analytical digestion and sequencing=== | ||
| + | |||
| + | '''Investigator: Clemens ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Confirm sucessful cloning | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed according to manufacturer's protocol (QIAprep, Qiagen) | ||
| + | * Analytic digestion was performed to check if the cloning was successful | ||
| + | |||
| + | * Batch for analytical digestion of P323/P324 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 0.5 µl | ||
| + | |Plasmid DNA P323/P324 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P325/P326 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 0.5 µl | ||
| + | |Plasmid DNA P326/P325 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P327/P328 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |1 µl/ 0.5 µl | ||
| + | |Plasmid DNA P328/P327 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P329/P330 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.5 µl/ 0.5 µl | ||
| + | |Plasmid DNA P329/P330 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P331/P332 with PstI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.5 µl/ 0.5 µl | ||
| + | |Plasmid DNA P331/P332 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for analytical digestion of P333/P334/P322 with AgeI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |0.5 µl/ 0.5 µl/ 0.5 µl | ||
| + | |Plasmid DNA P333/P334/P322 | ||
| + | |- | ||
| + | |1 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |EcoRI-HF (10 U/µl) | ||
| + | |- | ||
| + | |0.5 µl | ||
| + | |PstI-HF (20 U/µl) | ||
| + | |- | ||
| + | |7.5 µl/ 7.5 µl/ 7.5 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=10 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated for 1.5h at 37°C | ||
| + | * Samples were run on an 1% agarose gel for 45 min at 90V | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Except for P329, P330 and P322 all fragments were of the expected size | ||
| + | * Cloning of the inserts of P29 and P30 was started again. | ||
| + | |||
| + | For confirmation of the Sequence the following plasmids were send for sequencing: | ||
| + | * P323 (VF2 & VR) | ||
| + | * P331 (VF2) | ||
| + | * P333 (VF2) | ||
| + | * P328 (VF2) | ||
| + | * P326 (VF2) | ||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Preparative digestion of P333, P308, P331 and P301 === | ||
| + | |||
| + | '''Investigator: Clemens ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Cloning of new constructs | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | |||
| + | * Batch for preparative digestion of P333 with NgOMIV and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P333 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P333 with AgeI-HF and PstI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P333 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |AgeI-HF | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P308 with XbaI and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |15 µl | ||
| + | |Plasmid DNA P308 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |XbaI | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF | ||
| + | |- | ||
| + | |10 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P333 with SpeI-HF and EcoRI-HF | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |8 µl | ||
| + | |Plasmid DNA P331 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |EcoRI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |SpeI-HF | ||
| + | |- | ||
| + | |17 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Batch for preparative digestion of P301 with PstI-HF and NgOMIV | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''volume''' | ||
| + | |'''reagent''' | ||
| + | |- | ||
| + | |6 µl | ||
| + | |Plasmid DNA P301 | ||
| + | |- | ||
| + | |3 µl | ||
| + | |Cutsmart buffer (10x) | ||
| + | |- | ||
| + | |1 µl | ||
| + | |PstI-HF | ||
| + | |- | ||
| + | |1 µl | ||
| + | |NgOMIV | ||
| + | |- | ||
| + | |19 µl | ||
| + | |ddH2O | ||
| + | |- | ||
| + | |=30 µl | ||
| + | |'''TOTAL''' | ||
| + | |} | ||
| + | |||
| + | * Digestions were incubated for 3h at 37°C | ||
| + | * Samples were applied on a 1% agarose gel and run at 90V for 45min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * Digestions of P308, P331 and P301 showed the expected fragment sizes. | ||
| + | * These fragments were cut from the gel and purified by gelextraction (Promega) | ||
| + | * Samples were renamed and given Fragmentnumbers | ||
| + | |||
| + | * The Digestions of P333 showed fragments of unexpected length. The Ligation of eMA in pSB1C3 (previous step) failed. This ligation has to be repeated. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === PCR and purification of GSY7_new and GSY8 === | ||
| + | |||
| + | '''Investigator: MP, EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of GSY 7_new and GSY 8 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | *PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | * each GSY was amplified two times; once with Q5 GC-Enhancer and once without | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''25 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |5X Q5 Reaction Buffer | ||
| + | |5 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 mM dNTPs | ||
| + | |0.5 µL | ||
| + | |200 μM | ||
| + | |- | ||
| + | |10 μM Forward Primer (D20) | ||
| + | |1.25 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM Reverse Primer (VR) | ||
| + | |1.25 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |1 µL | ||
| + | |10 ng | ||
| + | |- | ||
| + | |Q5 High-Fidelity DNA Polymerase | ||
| + | |0.25 μl | ||
| + | |0.02 U/μl | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 25 μl (minus 5µl for GC-Enhancer respectively) | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||5 - 10 sec | ||
| + | |- | ||
| + | |61 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |16 °C | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Note: Use of the NEB T m Calculator is highly recommended. | ||
| + | |||
| + | Purification according to Promega PCR purification kit | ||
| + | |||
| + | '''Results:''' | ||
| + | no amplification GSY8 recognizable, GSY7did work | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Transformation of F271 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Transformation of F271 for further application | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA were added to blr cells | ||
| + | * incubation on ice for 20 min | ||
| + | * heatshock in 42°C for 40s | ||
| + | * incubation on ice for 4 min | ||
| + | * 700µl LB-medium were added | ||
| + | * incubation for 1h in 37°C shaker | ||
| + | * centrifugation for 3 min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Transformation of F1038 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Transformation of F1038 for further application | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA were added to blr cells | ||
| + | * incubation on ice for 20 min | ||
| + | * heatshock in 42°C for 40s | ||
| + | * incubation on ice for 4 min | ||
| + | * 700µl LB-medium were added | ||
| + | * incubation for 1h in 37°C shaker | ||
| + | * centrifugation for 3 min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting of eMA-HisTag === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * centrifugate at 6000 rpm for 30 min (OD= 6.15) | ||
| + | * 11,1 g pellet | ||
| + | * SDS-PAGE after protocol | ||
| + | * resuspend pellet in 20 mM Tris/HCl + 50 mM NaCl | ||
| + | * homogenize cells with PANDA | ||
| + | * centrifuge 30 min at 18000 rpm | ||
| + | * denaturate inclusion bodies in 16 ml Gnd-Chl (pH 1.5) over night | ||
| + | |||
| + | '''results:''' | ||
| + | |||
| + | [[File:Muc16_eMAHisTag1.jpg]] | ||
| + | |||
| + | marker, eMA before induction, eMA after induction, ULP after induction (SUMO protease) | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, Sep 2nd'''== | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === Ligation of F274+F275, F276+F260 and F279+F240 === | ||
| + | |||
| + | '''Investigator: ''' Clemens | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Cloning of new constructs | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting of eMA-HisTag === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * add 4 ml Gnd-Chl because pellet did not dissolve over night, continue shaking for few hours | ||
| + | * centrifugate for 15 min at 15000 rpm (protein in supernatant) | ||
| + | * refolding in 1.5 L cold PBS over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === purification of SUMO protease using ÄKTA purifier === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * centrifugation | ||
| + | * destroying DNA using ultrasonic for 10 min | ||
| + | * sterilfiltration | ||
| + | * purification due to HisTag affinity of protein to nickel in column (buffer: 20 mM Tris/HCl + 500 mM NaCl | ||
| + | * over night | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * purification stopped because of pressure alarm, was restarted an showed no SUMO protease in the sample | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === Cloning of extended MCS into pDSG and pcDNA5 vector === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Extending the MCS of pDSG and pcDNA5 for cloning of BioBricks | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Digestion of 5 µl P291 (pDSG) and 15 µl P339 (pcDNA) with SphI and NruI-HF for pDSG and MluI-HF for pcDNA5 | ||
| + | * Gelextraction of both signals at about 4 kb | ||
| + | * Oligo-hybridzation of O35/O36 and O36/O37 wo prior phosphorylation to obtain a total concentration of 17 ng/µl | ||
| + | * Ligation of oligo-hybridzation and vector-fragements according to calculations in the Inventarliste and overnight incubation at 16°C | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * expected signals after digestion: pDSG with 4 kb and 1.7 kb (lane 1), pcDNA with 4 kb and 1 kb (lane 2) | ||
| + | [[File:Muc16_160902_digestion_prep_gel_CC.jpeg|300px]] | ||
| + | |||
| + | </div> | ||
| + | <div class="general"> | ||
| + | === Miniprep of 2x pcDNA5 vector and 2x Streptavidin (nco) in pASK111 === | ||
| + | |||
| + | '''Investigator: CG''' | ||
| + | |||
| + | '''Aim of the experiment:''' Obtain plasmids for pcDNA5 vector and 2x Streptavidin (nco) in pASK111 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN) | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, Sep 3nd'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Transformation of F285; F286 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of F285; F286 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P1009 + PmeI&NotI === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P1009 + PmeI&NotI | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 33 µL DNA (~600 ng/µL) | ||
| + | * 2 µL PmeI | ||
| + | * 1 µL NotI | ||
| + | * 9 µL ddH2O | ||
| + | * 5 µL CutSmart buffer | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * F1039 80 ng/µL | ||
| + | </div> | ||
| + | |||
| + | =='''Sunday, Sep 4th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of F1040 (PCR product GSY7) === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Phosphorylation of F1040 (PCR product GSY7) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 15 µL F1040 (20 ng/µL) | ||
| + | * 5 µL T4 Ligase buffer | ||
| + | * 1 µL T4 PNK | ||
| + | * 29 µL ddH20 | ||
| + | * 30 min; 37 °C | ||
| + | * heat inactivation: 20 min; 65 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1041 (PCR product GSY7) | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Mung Bean nuclease (MBN) digestion of F1039 (pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 [PmeI+NotI]) === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Get rid of overhangs. Preparation of DNA for blunt ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 18 µL F1039 ( 80 ng/µL) | ||
| + | * 2 µL 10x CutSmart buffer | ||
| + | * 0.25 µL MBN (10 u/µL) | ||
| + | * 60 min; 30 °C | ||
| + | * purification by PCR purification kit, Promega | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1042 30 ng\µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F1042 +F1025 = F1043 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F1042 +F1025 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5.5 µL F1025 (9.2 ng\µL) | ||
| + | * 8.2 µL F1042 (35 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 3.5 µL ddH2O | ||
| + | * over night; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1042 +F1025 = F1043 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F1041 +F1029 = F1044 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F1041 +F1029 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1.6 µL F1029 (30 ng\µL) | ||
| + | * 23 µL F1041 (6 ng\µL) | ||
| + | * 3 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 1.3 µL ddH2O | ||
| + | * over night; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1041 +F1029 = F1044 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Cloning of SUMO-PAS in pASK75=== | ||
| + | |||
| + | '''Investigator: JL, EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | Ligation of F158 and "P310" (JL) (SUMO+PAS-Lysin in pASK75): | ||
| + | |||
| + | *5,6 µL | ||
| + | *2,4 µL | ||
| + | *2 µL | ||
| + | *1 µL T4-DNA-Ligase | ||
| + | *fill up with ddH2O to 20 µL | ||
| + | |||
| + | Transformation in NEB Turbo cells of 10 µL ligation | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =Week 17, Sep 5th - Sep 11th= | ||
| + | |||
| + | <div class="week" id="Week_17"> | ||
| + | |||
| + | =='''Monday, Sep 5th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting of eMA-HisTag === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * centrifugate after refolding at 11500 rpm for 30 min | ||
| + | |||
| + | '''results:''' | ||
| + | |||
| + | [[File:Muc16_eMAHisTag2.jpg]] | ||
| + | |||
| + | marker, eMA before induction, after induction, after denaturation and refolding | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Inocculation of P341 (E.coli Bl-21; 50 ml LB) in M9 medium for fermentation === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * M9 medium with Cam (1:1000) was inocculated with preculture (~50 mL) of P258 (pASK111+Streptavidin) | ||
| + | |||
| + | * the culture was incubated in the shaker over night at 37°C | ||
| + | |||
| + | '''results:''' | ||
| + | |||
| + | * a 50 ml culture with Ecoli Bl21 was induced | ||
| + | |||
| + | [[File:Muc16_erstgro.jpg]] | ||
| + | |||
| + | the SDS PAGE shows no streptavidin expression after induction | ||
| + | * next fermentation in pSA (not pASK111) | ||
| + | </div> | ||
| + | |||
| + | =='''Tuesday, Sep 6th'''== | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Phosphorylation of F1005, F1016 === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' to be completed | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation of F1045, F1046 === | ||
| + | |||
| + | '''Investigator: EF ''' | ||
| + | |||
| + | '''Aim of the experiment:''' to be completed | ||
| + | |||
| + | Ligation of F1005, F10016 (GelEx GSY8) in F1013 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Wednesday, Sep 7th'''== | ||
| + | |||
| + | <div class="general"> | ||
| + | |||
| + | === harvesting of eMA-HisTag === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' eMA-HisTag production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * precipitation with ammonium sulfate gradient (10% to 90%) | ||
| + | * the percentage was increased in 10% steps, after each addition of ammonium sulfate the solution was centrifugated at 8500 rpm for 20 min | ||
| + | * SDS-PAGE: samples of the supernatants | ||
| + | |||
| + | '''results:''' | ||
| + | |||
| + | [[File:Muc16_eMAHisTag3.jpg]] | ||
| + | |||
| + | The pellets of the precipitations with 50%, 60% and 70% were pooled and dialysed against IMAC buffer | ||
| + | Next step: Äkta | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="receptor"> | ||
| + | |||
| + | === MidiPrep of pSB1C3 with RFP generator (P4) === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' purification of CMV Nanoluc StrepTag PolyA (P312) | ||
| + | |||
| + | '''Procedure:''' | ||
| + | MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | 3268 ng/µl in 80 µl | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Inocculation of fermenter === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * inocculation with 50 ml preculture (P20 in LB medium) | ||
| + | * pO<sub>2</sub>= 60 %; Temp= 37°C, pH~7 | ||
| + | * growth over night | ||
| + | * OD next morning: 0.4 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, Sep 8th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === Fermentation === | ||
| + | |||
| + | '''Investigator:''' NA | ||
| + | |||
| + | '''Aim of the experiment:''' Streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * exponential feed (Amp) for ~13 h (about 3-4 L); start at OD= 7.5 (glucose test negative) | ||
| + | * Induction of streptavidin expression (IPTG) at OD= 30 | ||
| + | * production over night | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, Sep 9th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === harvesting streptavidin === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * OD= 41.4 | ||
| + | * centrifuge culture for 1 h at 4800 rpm | ||
| + | * resuspend pellet (275 g)in 20mM Tris HCl + 500 mM NaCl(pH 8) | ||
| + | * homogenize with PANDA | ||
| + | * the resulting lysate was transferred into centrifuge tubes and spun down (4°C, 9000 rpm, 30 mins). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight. | ||
| + | </div> | ||
| + | |||
| + | =='''Saturday, Sep 10th'''== | ||
| + | |||
| + | =='''Sunday, Sep 11th'''== | ||
| + | |||
| + | <div class="streptavidin"> | ||
| + | |||
| + | === harvesting streptavidin === | ||
| + | |||
| + | '''Investigator: NA''' | ||
| + | |||
| + | '''Aim of the experiment:''' streptavidin production | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * the denaturated protein solution was refolded in 15 L 1x PBS (added drop wise) over night | ||
| + | * the solution was centrifugated at 9500 rpm for 20 minutes, the pellet was stored in the freezer, the supernatant will be concentrated via cross flow and precipitated with ammonium sulfate afterwards | ||
| + | |||
| + | '''results:''' | ||
| + | |||
| + | [[File:Muc16_strepfermreinigung.jpg]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | |||
| + | =Week 18, Sep 12th - Sep 18th= | ||
| + | |||
| + | <div class="week" id="Week_18"> | ||
| + | =='''Tuesday, Sep 13th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P74 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P74 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | F1055: | ||
| + | |||
| + | * 2 µL DNA (1431 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL HincII (10 u/µL) | ||
| + | * 16 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | F1056: | ||
| + | |||
| + | * 2 µL DNA (1431 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL HincII (10 u/µL) | ||
| + | * 0.25 µL AgeI (20 u/µL) | ||
| + | * 15.5 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | F1057: | ||
| + | |||
| + | * 2 µL DNA (1431 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL HincII (10 u/µL) | ||
| + | * 0.25 µL NgoMIV | ||
| + | * 15.5 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_P74_div.png|250px]] | ||
| + | |||
| + | 1Kb ruler, F1055[HincII], F105[HincII&AgeI]6, F1057[HincII&NgoMIV], P1017[EcoRI&SpeI] | ||
| + | |||
| + | * F1055 49.5 ng/µL | ||
| + | * F1056 37.0 ng/µL | ||
| + | * F1057 29.7 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Analytical digestion of P1017 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Analytical digestion of P1017 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 2 µL DNA (312 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL EcoRI | ||
| + | * 15.5 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_P74_div.png|250px]] | ||
| + | |||
| + | 1Kb ruler, F1055[HincII], F105[HincII&AgeI]6, F1057[HincII&NgoMIV], P1017[EcoRI&SpeI] | ||
| + | |||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === PCR of GSY10 === | ||
| + | |||
| + | '''Investigator: MT''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | Amplification of GSY10 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''Component''' | ||
| + | |'''50 µL reaction''' | ||
| + | |'''Final concentration''' | ||
| + | |- | ||
| + | |Q5® High-Fidelity 2X Master Mix | ||
| + | |25 µL | ||
| + | |1 X | ||
| + | |- | ||
| + | |10 μM VR | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |10 μM VF2 | ||
| + | |2,5 μl | ||
| + | |0.5 μM | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |0.1 µL | ||
| + | |1 ng | ||
| + | |- | ||
| + | |Nuclease-Free Water | ||
| + | |to 50 μl | ||
| + | | | ||
| + | |} | ||
| + | |||
| + | Thermocycling Conditions for a Routine PCR: | ||
| + | |||
| + | {|cellspacing="0" border="1" | ||
| + | |'''STEP''' | ||
| + | |'''TEMP''' | ||
| + | |'''TIME''' | ||
| + | |- | ||
| + | |Initial Denaturation | ||
| + | |98 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |rowspan = "3"|35 cycles||98 °C||10 sec | ||
| + | |- | ||
| + | |62 °C | ||
| + | |30 sec | ||
| + | |- | ||
| + | |72 °C | ||
| + | |60 sec | ||
| + | |- | ||
| + | |Final extension | ||
| + | |72 °C | ||
| + | |120 sec | ||
| + | |- | ||
| + | |Hold | ||
| + | |4 °C | ||
| + | |- | ||
| + | |} | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Dephosphorylation of F1055 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Preparation of Vekor for blunt end Ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 15 µL F1055 (~50 ng\µL) | ||
| + | * 1 µL FastAP (1 u/µL) | ||
| + | * 2 µL 10x FastAP buffer | ||
| + | * 10 min ; 37 °C | ||
| + | * heat inactivation: 5 min; 75 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F348 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P1017 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P1017 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL DNA (312 ng/µL) | ||
| + | * 5 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 34.5 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p1017.png|250px]] | ||
| + | |||
| + | 1Kb ruler, P1017[SpeI&PstI], P1017[SpeI&PstI], F362: PCR-GSY7[AgeI], PCR-GSY7, PCR-GSY8 | ||
| + | |||
| + | * F350 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Over night liquid culture of P1009 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Getting enough cellmass for plasmid preparation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation | ||
| + | * Incubate over night at 37 °C in shaker | ||
| + | </div> | ||
| + | =='''Wednesday, Sep 14th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Gelextraction of F357[NgoMIV]; P1006[XbaI&PstI] === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Gelextaction of F357[NgoMIV]; P1006[XbaI&PstI] | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * Using gelextraction kit, Promega | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Muc16_gelex_F364_F365.png|250px]] | ||
| + | |||
| + | 1kb ladder, F357[NgoMIV]; P1006[XbaI&PstI] | ||
| + | |||
| + | * F357[NgoMIV] -> F364 41 ng/µL | ||
| + | * P1006[XbaI&PstI] -> F365 9.1 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation: F1057 + F364 = F366 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F1057 + F364 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1,6 µL F1057 (29.7 ng\µL) | ||
| + | * 2.1 µL F364 (41 ng\µL) | ||
| + | * 3 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 22.3 µL ddH2O | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F1057 + F364 = F366 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F350 + F365 = F367 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F350 + F365 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2,6 µL F350 (19 ng\µL) | ||
| + | * 2.1 µL F365 (9.1 ng\µL) | ||
| + | * 3 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 15.4 µL ddH2O | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F350 + F365 = F367 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P1008 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P1008 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL P1008 (256.5 ng/µL) | ||
| + | * 5 µL CutSmart buffer | ||
| + | * 0.25 µL XbaI | ||
| + | * 0.25 µL PstI | ||
| + | * 34.5 µL ddH2O | ||
| + | * incubation for 1h at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p1008_2.png|250px]] | ||
| + | |||
| + | 1Kb ruler, ... P1008[XbaI&PstI] | ||
| + | |||
| + | * P1008 -> F368 ~1 ng/µL | ||
| + | |||
| + | * Maybe do it again, fragments do not have the right size, apparently. | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F366; F367 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Amplification of Vector | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * Transformation following SOP(13.02.2013) | ||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | |||
| + | === Analytical digestion of P296 and F269 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Analyze construct for eventual preparative digestion | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 2 µl DNA | ||
| + | 15µl ddH2O | ||
| + | 1µl of PstI | ||
| + | 2µl cut smart buffer | ||
| + | |||
| + | incubation at 37°C over night | ||
| + | Gel electrophoresis and gel extraction | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | === Preparative digestion of P313, P296 and P73 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Prepare construct for ligation later on | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | 10 µl (P73)/ 15 µl (P296)/ 20 µl (P313) DNA | ||
| + | 2µl of each enzyme (P296&P313 --> EcoRI&SpeI, P73 --> EcoRI&XbaI) | ||
| + | 5µl cut smart buffer | ||
| + | ad 50 µl ddH2O | ||
| + | |||
| + | incubation at 37°C over night | ||
| + | Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | * P296 [EcoRI&SpeI] --> F378 | ||
| + | * P313 [EcoRI&SpeI] --> F383 | ||
| + | * P73 [EcoRI&XbaI] --> F379 | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | === Ligation: F269 + F270 = F380; F269 + F92 = F381; F269 + F133 = F382 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Ligation: | ||
| + | |||
| + | ** F269 + F270 = F380 | ||
| + | ** F269 + F92 = F381 | ||
| + | ** F269 + F133 = F382 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL F269 (6,3 ng) | ||
| + | * 0,5 µL F270 (5,6ng)/ 0,25 µl F92 (6,75 ng)/ 0,25 µl F133 (13 ng) | ||
| + | * 0,5 µL buffer | ||
| + | * 0,5 µL T4 ligase | ||
| + | * 0,5/0,75 µL ddH2O | ||
| + | * ONT; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F269 + F270 = F380 | ||
| + | * F269 + F92 = F381 | ||
| + | * F269 + F133 = F382 | ||
| + | </div> | ||
| + | |||
| + | <div class="kill-switch"> | ||
| + | === Sequencing of P313 === | ||
| + | |||
| + | '''Investigator: EU''' | ||
| + | |||
| + | '''Aim of the experiment:''' Sequencing of P313 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | *Sequencing batch was prepared after manufacturer's protocol: | ||
| + | |||
| + | **6 µl of plasmid DNA (end concentration 58,6 ng) in 9µl H2O | ||
| + | **2 µl sequencing primer (VF2) | ||
| + | |||
| + | *The different vectors we sequenced received the following barcodes: | ||
| + | |||
| + | ** P313 with VF2: FR12904387 | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Thursday, Sep 15th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P1008 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P1008 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL P1008 (256.5 ng/µL) | ||
| + | * 5 µL CutSmart buffer | ||
| + | * 0.25 µL XbaI | ||
| + | * 0.25 µL PstI | ||
| + | * 34.5 µL ddH2O | ||
| + | * incubation for over night at 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p1008_3.png|250px]] | ||
| + | |||
| + | P1008[XbaI&PstI], P1008[XbaI&PstI], 1Kb ruler | ||
| + | |||
| + | * P1008[XbaI&PstI] -> F375 32 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Ligation: F63 + F375 = F377 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F63 + F375 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2.2 µL F63 (45 ng\µL) | ||
| + | * 6.3 µL F375 (32 ng\µL) | ||
| + | * 3 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 17.5 µL ddH2O | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F63 + F375 = F377 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Fill in of overhangs of F386 with Large Klenow fragment === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * 3' overhang removal and fill-in of 3' recessed (5' overhang) ends using DNA Polymerase I, Large (Klenow) Fragment | ||
| + | * short: create blunt ends, needed for ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 17.09 µL F386 (25.9 µL) | ||
| + | * 0.66 µL 10 mM dNTP Mix | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 0.25 µL Klenow Fragment | ||
| + | * 15 min; RT | ||
| + | * heat inactivation: 20 min; 75 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F387 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Incubation of F386 with Q5 polymerase === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Create blunt ends, needed for ligation | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 5 µL 2x Q5 Mastermix | ||
| + | * 5 µL F386 (25.9 ng/µL) | ||
| + | * 15 min; 72 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F388 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F374 + F386 = F390 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F374 + F386 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 1 µL F374 (25.9 ng\µL) | ||
| + | * 17.5 µL F387 (~ 22 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F374 + F386 = F390 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F374 + F388 = F391 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F374 + F388 = F391 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 0.3 µL F374 (25.9 ng\µL) | ||
| + | * 10 µL F388 (~ 13 ng\µL) | ||
| + | * 2 µL T4 ligase buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 6.7 µL ddH2O | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F374 + F388 = F391 | ||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | === Transformation of F390, F391, F376 and F377 (INSULIN) === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Transformation of F390, F391, F376 and F377 for further application | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA were added to xl1-cells | ||
| + | * incubation on ice for 20 min | ||
| + | * heatshock in 42°C for 40s | ||
| + | * incubation on ice for 4 min | ||
| + | * 700µl LB-medium were added | ||
| + | * incubation for 1h in 37°C shaker | ||
| + | * centrifugation for 3 min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F389 (zk-vector) === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Transformation of F389 for further application | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA were added to xl1-cells | ||
| + | * incubation on ice for 20 min | ||
| + | * heatshock in 42°C for 40s | ||
| + | * incubation on ice for 4 min | ||
| + | * 700µl LB-medium were added | ||
| + | * incubation for 1h in 37°C shaker | ||
| + | * centrifugation for 3 min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="hypoxia"> | ||
| + | |||
| + | === Transformation of F366 === | ||
| + | |||
| + | '''Investigator: EU ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | * Transformation of F366 for further application | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3 µL DNA were added to xl1-cells | ||
| + | * incubation on ice for 20 min | ||
| + | * heatshock in 42°C for 40s | ||
| + | * incubation on ice for 4 min | ||
| + | * 700µl LB-medium were added | ||
| + | * incubation for 1h in 37°C shaker | ||
| + | * centrifugation for 3 min | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | </div> | ||
| + | |||
| + | =='''Friday, Sep 16th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P430 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P430 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL 430 (~150 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL PacI | ||
| + | * 0.25 µL AgeI | ||
| + | * 7.5 µL ddH2O | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p430_etc.png|250px]] | ||
| + | |||
| + | 1kb ruler; P430[PacI&AgeI], P424[SpeI&PstI], P425[SpeI&PstI] | ||
| + | |||
| + | * P430[PacI&AgeI] -> F400 2.1 ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P424 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P424 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 17 µL 424 (60.9 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p430_etc.png|250px]] | ||
| + | |||
| + | 1kb ruler; P430[PacI&AgeI], P424[SpeI&PstI], P425[SpeI&PstI] | ||
| + | |||
| + | * P424[SpeI&PstI] -> F401 15.7ng/µL | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P425 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P425 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | * 10 µL 425 (~222 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 7.5 µL ddH2O | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_p430_etc.png|250px]] | ||
| + | |||
| + | 1kb ruler; P430[PacI&AgeI], P424[SpeI&PstI], P425[SpeI&PstI] | ||
| + | |||
| + | * P425[SpeI&PstI] -> F402 53 ng/µL | ||
| + | </div> | ||
| + | </div> | ||
| + | =Week 19, Sep 19th - Sep 25th= | ||
| + | <div class="week" id="Week_19"> | ||
| + | =='''Tuesday, Sep 20th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Digestion of P444; P445; P446 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | * Digestion of P444; P445; P446 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | |||
| + | P444: | ||
| + | * 8 µL P444 (377.7 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 9.5 µL ddH2O | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | P445: | ||
| + | * 14 µL P445 (223.7 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 3.5 µL ddH2O | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | P446: | ||
| + | * 17.5 µL P446 (144 ng/µL) | ||
| + | * 2 µL CutSmart buffer | ||
| + | * 0.25 µL SpeI | ||
| + | * 0.25 µL PstI | ||
| + | * 1h; 37 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | |||
| + | [[File:Muc16_gelextraction_P451_p4444_P446_etc.png|250px]] | ||
| + | |||
| + | 1kb ruler P451[PacI&MluI], P451[PacI&MluI], P444[SpeI&PstI], P446[SpeI&PstI] | ||
| + | |||
| + | -> I did P444 again, because there was marker in the line. | ||
| + | |||
| + | [[File:Muc16_gelextraction_P451_p4444_P445_etc.png|250px]] | ||
| + | |||
| + | 1kb ruler, P444[SpeI&PstI], P445[SpeI&PstI] | ||
| + | |||
| + | * P444[SpeI&PstI] -> F420 18 ng/µL | ||
| + | * P445[SpeI&PstI] -> F418 24.2 ng/µL | ||
| + | * P446[SpeI&PstI] -> F421 7.4 ng/µL | ||
| + | </div> | ||
| + | =='''Wednesday, Sep 21th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation {F438 + F435} + F433 or F334 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation {F438 + F435} + F433 or F334 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | |||
| + | F438 + F435: | ||
| + | |||
| + | * 10 µL F438 (40.2 ng/µL) | ||
| + | * 11 µL F435 (30.5) | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 1 µL T4 Ligase | ||
| + | * 6 µL ddH2O | ||
| + | |||
| + | {F438 + F435} + F433: | ||
| + | |||
| + | * 0.5 µL F433 (48.2 ng/µL) | ||
| + | * 4.5 µL {F438 + F435} (~24.4 ng/µL) | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 1 µL T4 Ligase | ||
| + | * 12 µL ddH2O | ||
| + | |||
| + | {F438 + F435} + F434: | ||
| + | |||
| + | * 0.5 µL F434 (109 ng/µL) | ||
| + | * 4.5 µL {F438 + F435} (~24.4 ng/µL) | ||
| + | * 2 µL T4 Ligase buffer | ||
| + | * 1 µL T4 Ligase | ||
| + | * 11.5 µL ddH2O | ||
| + | |||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | * F438 + F435 = F439 | ||
| + | * {F438 + F435} + F434 = F440 | ||
| + | * {F438 + F435} + F433 = F441 | ||
| + | </div> | ||
| + | |||
| + | =='''Sunday, Sep 25th'''== | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F457 + F463 = F464 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F457 + F463 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 2.7 µL F457 (29.3 ng\µL) | ||
| + | * 4.3 µL F463 (22.1 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F457 + F463 = F464 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F458 + F463 = F465 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F458 + F463 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 3.8 µL F458 (15.5 ng\µL) | ||
| + | * 3.2 µL F463 (22.1 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * 30 min; RT | ||
| + | |||
| + | '''Results:''' | ||
| + | * F458 + F463 = F465 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F434 + F464 = F466 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F434 + F464 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 0.5 µL F434 (109.1 ng\µL) | ||
| + | * 6.5 µL F464 (17.4 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * over night; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F434 + F464 = F466 | ||
| + | </div> | ||
| + | <div class="hypoxia"> | ||
| + | === Ligation: F434 + F465 = F467 === | ||
| + | |||
| + | '''Investigator: MT ''' | ||
| + | |||
| + | '''Aim of the experiment:''' | ||
| + | |||
| + | *Ligation: F434 + F465 | ||
| + | |||
| + | '''Procedure:''' | ||
| + | * 0.4 µL F434 (109.1 ng\µL) | ||
| + | * 6.6 µL F465 (12.96 ng\µL) | ||
| + | * 2 µL buffer | ||
| + | * 1 µL T4 ligase | ||
| + | * over night; 16 °C | ||
| + | |||
| + | '''Results:''' | ||
| + | * F434 + F465 = F467 | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | =Week 20, Sep 26th - Aug 2th= | ||
| + | <div class="week" id="Week_20"> | ||
| + | |||
| + | </div> | ||
Latest revision as of 03:14, 20 October 2016
Labjournal
Samples
Transformation of E. coli XL1 blue with Phytochrome B (2–908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion
Procedure:
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker
- 100 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with
Investigator:
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
Week 1, May 16th - May 22nd
Monday, May 16th
Streptavidin Plasmid Check
Investigator: JB, LK, JH
Aim of the experiment: Verify cloning
Procedure:
- MiniPrep following manufacturer's protocol (QIAprep MiniPrep, Qiagen) (4 clones each of pSA1, pSAm1 in pASK75)
- Analytic digestion with 0.25 µL XbaI, 0.25 µL HindIII (HF), 1 µL SmartCut Buffer, 5 µL plasmid DNA, 3.5 µl H2O
- 5 µL analyzed in gel electrophoresis (1% agarose)
Results: Successful cloning verified. Stored at –20 °C
- Lane 1: 5 µL Thermo Fisher, 1 kb ladder
- Lanes 2–9: 5 µL digestions of P6–P13, band of SA (mut1) at about 300 bp, band of digested plasmid at about 3,000 bp
Streptavidin Expression: Transform BL21
Investigator: JB, JH
Aim of the experiment: Express pSA1 and pSAm1 in E. coli BL21
Procedure:
- Transformation into competent E. coli BL21 according to protocol of P6 and P10
Results: Plates (LB Amp) in incubator (37 °C) for further processing
Tuesday, May 17th
SDS Gel Analysis
Investigator: CG
Aim of the experiment: Analyze collagen 1/2, eGFP, fraction 30 of egg precipitation via SDS gel
Procedure:
- Mixed 80 µL samples with 20 µL SDS buffer
- Heated at 95 °C for 10 min
- 1 d staining, 1 d unstaining
Results: Successful cloning verified. Stored at –20 °C
- Lane 1: 8 µL marker (Thermo Fisher #26610)
- Lane 2: Fraction 30 (IEC), 3 µL, band at 35 kDa, avidin expected at 16 kDa
- Lane 3: eGFP, 12 µL, band at 27 kDa, eGFP expected at 27 kDa, many impurities
- Lane 4: Collagen 1, 12 µL, no sharp band
- Lane 5: Collagen 1, 12 µL, no sharp band
MiniPreps pSb1C3–AviTag, –A3C5 and pASK75–(SA1), –(SAm1)
Investigator: CR, CG
Aim of the experiment: Verify cloning
Procedure:
- MiniPrep was performed following manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Analytic digestion with 0.25 µL XbaI, 0.25 µL HindIII (HF) for pASK plasmids and 0.25 µL NgomIV, 0.25 µL AgeI (HF) for pSb1C3 plasmids, 1 µL SmartCut Buffer, 5 µL plasmid DNA, 3.5 µL H2O
- 5 µL on 1% agarose gel electrophoresis
Results: Successful cloning verified for pASK plasmids. Repeat pSb1C3 plasmids. Stored at –20 °C
- Lane 1: 5 µL Thermo Fisher, 1 kb Ladder
- Lane 2: 5 µL digestion of pSb1C3–AviTag
- Lane 3: 5 µL digestion of pSb1C3–A3C5
- Lane 5: 5 µL digestion of pASK75(SA1), EB elution
- Lane 6: 5 µL digestion of pASK75(SAmut1), EB elution
- Lane 7: 5 µL digestion of pASK75(SAmut1), H2O elution
Inoculation of Pre-Culture with BL21 (pASK75 (SA1)) in LB Medium
Investigator: CR
Aim of the experiment: Pre-culture for streptavidin expression in TB medium
Procedure:
- Added 50 µL ampicillin to 50 mL LB medium
- Picked colonies from BL21 (pASK75 (SA1))
- Inoculated LB medium
- Incubated at 30 °C overnight
Wednesday, May 18th
Repeat Analytical Gel of pSb1C3–AviTag, –A3C5
Investigator: CG, CR
Aim of the experiment: Verify cloning
Procedure:
- Analytic digestion with 0.25 µL NgoMIV, 0.25 µL AgeI (HF) for pSb1C3 plasmids, 1 µL SmartCut Buffer, 8.5 µL plasmid DNA
- 10 µL analyzed in gel electrophoresis (2% agarose)
Results:
Inoculation of BL21 (pASK75 (SA1)) Culture in 2 L TB Medium and Induction of Streptavidin Production by Addition of Tetracycline
Investigator: CR
Aim of the experiment: Produce streptavidin
Procedure:
- Ampicillin (2 mL) was added to the medium (1:1000)
- The pre-culture (50 mL) was poured into the medium
- Culture incubated at 37 °C and 140 rpm until OD550 reached 0.5
- To induce streptavidin expression, anhydro-tetracycline (200 µL) was added to the culture (1:10,000)
- The culture was incubated at 37 °C and 140 rpm for 4 hours
Results: Streptavidin expression by BL21
Expression and Harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB Medium
Investigator: CR, JB, JH
Aim of the experiment: Recombinant expression and purification of streptavidin
Procedure:
- After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor)
- The supernatant was discarded, and the pellet was transferred into a beaker of sufficient size and resuspended in Tris Buffer B cooled to 4 °C (50 mM Tris/HCl pH = 8.0, 1 mM EDTA)
- The solution was homogenized in the PANDA (ask supervisor)
- The resulting lysate was transferred into centrifuge tubes and spun down (4 °C, 18,000 rpm, 10 min, XX34-rotor). The supernatant was discarded, and the pellet was resuspended in 6 M Gua-HCl (pH = 1.5) at 4 °C overnight.
Dialysis of eGFP
Investigator: NA, JH, CR
Aim of the experiment: Purify eGFP
Procedure:
- eGFP was thawed on ice
- eGFP was then poured into a dialysis hose (cut-off 14 kDa)
- The hose was then placed in ice-cold Tris/HCl 20 mM pH 8.0
- Dialyzed at 4 °C overnight (XX34 rotor). The supernatant was discarded, and the pellet was
MiniPrep of quickchanged pNGAL146-A2 EspP
Investigator: NA
Aim of the experiment: Extraction of pNGAL146-A2 plasmid from XL1 blue
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
Sequencing of P14 (Avi-Tag), P15 (A3C5) & P19 (Quick change (QC) EspP in pNGAL-A2)
Investigator: CR, NA
Aim of the experiment: Sequencing of P14 (Avi-Tag), P15 (A3C5) & P19 (Quick change (QC) EspP in pNGAL-A2)
Procedure:
- Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
- The different plasmids we prepared received the following barcodes:
- P14 : FR11326653
- P15 : FR11326655
- P19 (K4): FR11326654
- P16 (K1): FR11326652
- P17 (K2): FR11326651
- P18 (K3): FR11326650
Digestion of P16 , P17, P18 & P19 (all QC EspP in pNGAL-A2) with AgeI & HindIII + analytical gel
Investigator: NA, JH, CR
Aim of the experiment: Verification of success of quickchange
Procedure:
- analytic digestion with: 0,25 µl HindIII (HF), 0,25 µl AgeI (HF), 1 µl SmartCut Buffer, 500 ng plasmid-DNA, fill up with ddH2O (Vtotal= 10µL)
- 10 µl on 2% agarose gel for electrophoresis
Results: No signal at 600 bp --> quickchange seems to be successful (waiting for sequencing)
Thursday, May 19th
Re-Sequencing of P19
Investigator: CR
Aim of the experiment: Re-Sequencing of P19
Procedure:Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- P19 (K4): FR11326649
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CG
Aim of the experiment: Re-Trafo of pSB1C3 RFP for later on: digestion, dephosphorylation and cloning
Procedure: transformation according to protocol of P4 E. Coli XL1
Result: plates (LB Cam) in incubator for further processing (37 °C)
Streptavidin refolding
Investigator: JB
Aim of the experiment: Refolding of denaturated Streptavidin
Procedure: After the pellet had almost completely dissolved in 6M GdmCl, the solution was spun down (4°C, 20 mins, 18,000 rpm). The supernatant was transferred carefully into a falcon tube and the pellet was cast away. Via a hydraulic pump (flow rate: 2x10 ml/min) the lysate was transferred Into 5L PBS 1x. Afterwards the pump was cleaned with technical isopropanol and ELGA water. The solution was stirred overnight at 4°C for refolding.
Biotinylation of BSA
Investigator: JB
Aim of the experiment: Biotinylation of BSA
Procedure: A 100 µM (=6.8 mg/ml) solution of BSA (Albumin fraction V, pH=7, in the fridge in the central lab) was created (V=10 ml). 220 µl of a 100 mM Biotin stock were added. The mixture was stored overnight Iin the fridge (4°C).
Result: Hopefully biotinylated BSA mixture in the fridge (4°C).
Friday, May 20th
Qualitative analysis of streptavidin expression
Investigator: CG
Aim of the experiment: SDS gel analysis of recombinant strepatividin expression
Procedure: mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining
Result:
- 1. Lane: 8 µl Marker (Thermo Fisher #26610)
- 2. Lane: 12 µl culture aliquot, before induction
- 3. Lane: 12 µl culture aliquot, after induction
- 4. Lane: 3 µl culture aliquot of the lysed pelet, Strepatvidin expected at about 16 kDa
- 5. Lane: 3 µl culture aliquot of the supernatend after lysis, no Strepatvidin expected at about 16 kDa
- 6. Lane: 1.5 µl culture aliquot of the lysed pelet, Strepatvidin expected at about 16 kDa
- 7. Lane: 1.5 µl culture aliquot of the supernatend after lysis, no Streptavidin expected at about 16 kDa
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: LK
Aim of the experiment: Inoculation of pre-culture with E. coli XL1 (pSb1C3 -RFP) in LB-Chloramphenicol-medium
Procedure: Picking of colonies for E. coli XL1 (pSb1C3 -RFP) Inoculate in 5 ml LB-Chloramphenicol-medium Incubate at 37°C over night at 200 rpm
Transformation of Biobricks in XL1-blue
Investigator: NA, JH
Aim of the experiment: Transformation
Procedure: -10 µl dd H2O in well of interest (standard distribution kit) -1 µl Plasmid (out of well) to cells -Transformation according to the SOP Used bricks: K577893, B0015, R0040, B0032, I14033, K747096
Ammonium sulfate precipitation of streptavidin
Investigator: JB
Aim of the experiment: Reduction of the protein solution volume and precipitation of streptavidin
Procedure: The 5 L protein solution was spun down (20 mins, 5,000 rpm) and the supernatant transferred into a beaker. In order to lower the volume of the solution for ammonium sulfate precipitation, the solution was first filtered via a membrane crossflow pump (membrane: Sartocon 0.45 µm, thick membrane).
Dialysis of biotinylated BSA
Investigator: NA
Aim of the experiment: purification of biotinylated BSA
Procedure: dialysis against Tris/HCl 20 mM, pH 8, 10 mM NaCl over night; cut off : 14 kDa
Week 2 (May 23rd - May 29th)
Monday, May 23th
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CR
Aim of the experiment: Cloning of A3C5 and Avi-Tag into pSB1C3
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- pSB1C3 was digested with AgeI and NgOMIV (10 µL plasmid + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 14 µL ddH2O; incubation for 1h, 37°C)
- FastAP was added and the reaction was incubated for another 2h at 37°C
- The digestion was purified by gelelctrophoresis (1%, 75V, 1h)
- Plasmid backbone was cut from the gel
Results:
Signal 1: linearized pSB1C3 (successfull digestion)
Signal 2: supercoiled pSB1C3 (digestion not successfull)
Signal 3: RFP-generator
--> next time gel should run longer (just if you need the back bone)
Filtration of Streptavidin and precipitation with Ammonium-sulfate
Investigator: MP, CR
Aim of the experiment: Concentration of Streptavidin
Procedure: The solution was filtered via a membrane crossflow pump (membrane: Sartocon 0.45 µm, thick membrane). The final volum was 0.5L The solution was precipitated by addition of Ammonium-sulfate (2 steps: 1. 40% Ammonium-sulfate; 2. 70% Ammonium-sulfate) After the first addition of ammonium-sulfate the solution was stirred (1h, 4°C) Afterwards the solution was centrifuged (10.000 rpm; 30 min) and the supernatant was used for the second precipitation step The solution was stirred again (over night, 4°C)
Inoculation of pre-culture with BioBricks in pSB1C3 in LB-Cam
Investigator: JH, JL
Aim of the experiment:
Procedure: Add 50 µL chloramphenicol in 50 mL LB-medium Picking colonies from K577893; B0015; B0032 (in pSB1C3) * Inoculate 4 mL medium (CAM) Incubate at 37°C over night
- Following cultures didn´t grow and were therefore not inoculated:
R0040; K747096; I14033
Retransformation of Biobricks in XL1-blue
Investigator: JH, JL, EF
Aim of the experiment: Transformation
Procedure: 10 µl dd H2O in well of interest (standard distribution kit) 1 µl Plasmid (out of well) to cells Transformation according to the SOP Used bricks: K577893, B0015, R0040, B0032, I14033, K747096
Tuesday, May 24th
Inoculation of pre-culture with BioBricks in pSB1C3 in LB-Cam
Investigator: CR
Aim of the experiment:
Procedure: Add 50 µL chloramphenicol in 50 mL LB-medium Picking colonies from K577893; B0015; B0032 (in pSB1C3) * Inoculate 4 mL medium (CAM) Incubate at 37°C over night
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CR
Aim of the experiment: Gelextraction of Signal 1 from pSB1C3-digestion
Procedure: Gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen)
Ammonium sulfate precipitation
Investigator: JB
Aim of the experiment: Precipitation the 70% saturated ammonium sulfate solution
Procedure: The ammonium sulfate solution, which actually was a suspension, was transferred into centrifuge tubes and spun down (10,000 rpm, 45 mins). As it turns out, unfortunately too much ammonium sulfate was used for the precipitation (wrong table). The precipitate was brought back into solution (20 mM Tris/HCl, pH 8.0) and loaded onto an SDS gel for analysis, along with other samples (flowthrough of the filtration from the day before).
MiniPrep of BioBricks in pSB1C3 (inoculated on May 23th)
Investigator: JH
Aim of the experiment: Extraction of BioBricks B0032, B0015 and K577893 in pSB1C3 from XL1 blue
Procedure: MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) concentrations measured (in ng/µL): (B0032) 360.2; (B0015) 254.9; (K577893) 99.2
Sequencing of P23 (weak RBS), P24 (double terminator) & P24 (Tet repressed GFP)
Investigator: JH
Aim of the experiment: Sequencing of P23, P24 & P25 (x2)
Procedure: Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- P23 (VF2) : FR11326647
- P24 (VF2) : FR11326646
- P25 (VF2) : FR11326645
- P25 (VR) : FR11326644
Wednesday, May 25th
Inoculation of pre-culture with tet-repressed GFP (P25) in LB-Cam
Investigator: NA
Procedure: Add 50 µL chloramphenicol to 50 mL LB-medium Picking colonies from plate (BL21) Incubate over night at 37°C => dismissed, because plasmid sequence was inaccurate
MiniPrep of K747096, I14033, R0040, B0015, K577893 and B0032
Investigator: CR
Aim of the experiment: MiniPrep of BioBricks
Procedure: MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) Concentrations were measured (in ng/µL):
- K747096: 286.2
- I14033: 246.1
- R0040: 243.5
- B0015: 244.1
- K577893: 440.9
- B0032: 259.7
PCR of P19 (QC EspP) with O3 and O4
Investigator: CR
Aim of the experiment: Amplification of quickchanged EspP
Procedure:
- reaction was performed after manufacturer's protocol (PCR Using Q5® High-Fidelity DNA Polymerase, NEB)
- 274,4 pg Plasmid were used for amplification (1 µL)
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial Denaturation | 98 | 30 |
| 35 cycles | 98 | 10 |
| 72 | 30 | |
| 72 | 60 | |
| final Extension | 72 | 5 |
| Hold | 4 | ꝏ |
- PCR product was purified accoding to manufacturer's protocol (PCR puification kit, Qiagen)
- DNA was eluted 40 µL EB buffer
Digestion of PCR of EspP (P19 with O3 and O4)
Investigator: CR
Aim of the experiment: Digestion of EspP with AgeI and NgoMIV to ligate it in pSB1C3 later on
Procedure:
- Batch for preparative digestion:
| Volume | Reagent |
| 38.5 µL | PCR product P19 with O3 and O4 |
| 1 µL | AgeI |
| 1 µL | NgoMIV |
| 5 µL | CutSmart buffer |
| 4.5 µL | ddH2O |
| 50 µL | TOTAL |
- Incubation for 2 h at 37 °C
Sequencing of P26 (CMV promotor), P27 (p cat promotor), P28 (Tet repressible promotor), P30 (Tet repressed GFP)
Investigator: NA
Aim of the experiment: Sequencing of P26, P27, P28 & P30 (x2)
Procedure: Sequencing batches were prepared after manufacturer's protocol (15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer) The different plasmids we prepared received the following barcodes: P26 (VF2) : FR11326643 P27 (VF2) : FR11326642 P28 (VF2) : FR11326641 P30 (VF2) : FR11326640 P30 (VR): FR11326639
Ion exchange chromatography of eGFP
Investigator: NA, JB,JL
Aim of the experiment: purification of eGFP
Procedure:
- use Äkta-Purifier (Q-column for eGFP (quarternary ammonium as stationary phase)). Ask Andy before use!
- Pump A: running buffer (20 mM Tris/HCl pH 8); pump B: elution buffer (20 mM Tris/HCl pH 8 and 1,000 mM (1 M) NaCl
- basic pumpwash before start
- injection in loop
- gradient of ion strength on column.
Results: Elution of eGFP at ~200 mM NaCl.
Thursday, May 26th
Inoculation of cultures with XL-1 F11 and F12 clones
Investigator: JB
Aim of the experiment: Inoculation of a 5 ml culture with a clone of the previously transformed F11 (Avi-Tag) F12 (Antibody-binding site).
Cloning of ligation F14 +F13
Investigator: MP, EF, JH
Aim of the experiment: new biobrick EspP in pSB1C3
Procedure: F14 was purified by gelelctrophoresis (1%, 90 V, 40 min) gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen) ligation of F14 (EspP) with F13 (dephos. PSB1C3) transformation according to protocol in competent E.coli XL1 blue
Results:
Transformation of Biobrick B0034 in XL1-blue
Investigator: JH
Aim of the experiment: Transformation
Procedure:
- 10 µl ddH2O in well of interest (standard distribution kit, plate 4, well 1N)
- 1 µl Plasmid (out of well) to cells
- Transformation according to the SOP (incl. rescue)
Transformation of Biobricks K577895 and K577894 in XL1-blue
Investigator: NA
Aim of the experiment: Transformation
Procedure:
- 10 µl ddH2O in well of interest (standard distribution kit, plate 1, wells 14B / 12P)
- 1 µl Plasmid (out of well) to cells
- Transformation according to the SOP (incl. rescue)
SDS-PAGE of eGFP-fractions after IEC
Investigator: NA, CG
Procedure: mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining Concentrations were measured via nanodrop (A280, extinction coefficient: 55000 M−1cm−1)
Result: concentrations in mg/ml 1. Lane: 8 µl Marker 2. Lane: 10 µl of fraction 10; c= 0 3. Lane: 10 µl of fraction 11; c= 0,15 4. Lane: 10 µl of fraction 12; c= 0,77 5. Lane: 10 µl of fraction 13; c= 0,29 6. Lane: 10 µl of fraction 14; c= 0,4 7. Lane: 10 µl of fraction 15; c= 0,144 8. Lane: 10 µl of fraction 16; c= 0,085
eGFP is clearly detectable at about 27 kDa. The most pure fractions 12 and 13 were pooled for later biotinylation.
Friday, May 27th
Inoculation of pre-cultures with B0034 (strong RBS), K577894 (Tet repressed YFP), KK577895 (Tet repressed RFP), F13+F14
Investigator: JH
Procedure:
- Add 50 µL ampicillin(B0034)/chloramphenicol(K5777894, K577895, F13+F14) to 50 mL LB-medium
- Picking colonies from plate (XL1 blue; all rescue)
- Incubate over night at 37°C
Inoculation of BL21 (pASK75 (SA1)) culture in 2 L TB-Medium and induction of streptavidin production by addition of tetracycline
Investigator: NA
Aim of the experiment: Production of streptavidin
Procedure:
- Ampicillin (2 mL) was added to the Medium (1:1000)
- The pre-culture (50 mL) was poured into the Medium
- Culture was incubated at 37°C and 140 rpm until OD550 reached 0.5
- To induce streptavidin expression anhydro-tetrazycline (200 µL) was added to the culture (1:10000)
- The culture was incubated at 37°C and 140 rpm for 4 hours
Result: Streptavidin expression by BL21
Re-sequencing of P30 (Tet repressed GFP in pSB1C3)
Investigator: NA
Aim of the experiment: Purification and sequencing of P30
Procedure:
- P30 was heated up to 100 °C and centrifugated afterwards to remove proteins from Promotor (if there are some), the supernatant is sequenced
- Sequencing batches were prepared after manufacturer's protocol
- (15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer)
- The different plasmids we prepared received the following barcodes:
- P30 (VF2) : FR11326638
Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium
Investigator: JH, NA
Aim of the experiment: Recombinant expression and purification of Streptavidin
Procedure:
- After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4°C, 5000 rpm, 20 mins, F 4X1L rotor)
- The supernatant was cast away and the pellet was stored at –20 °C
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CG
Aim of the experiment: MiniPrep of cloned pSB1C3-A3C5 and –AviTag Plasmids
Procedure:
- MiniPrep of inoculated precultures was performed according to manufacturer’s protocol (QIAprep MiniPrep, Qiagen)
- Concentrations were measured (in ng/µL):
Colony 1 of pSB1C3- AviTag P32: 120 Colony 2 of pSB1C3- AviTag P33: 110 Colony 1 of pSB1C3-A3C5 P34: 126 Colony 2 of pSB1C3-A3C5 P35: 188
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: NA, JH, CG
Aim of the experiment: Initial vector digestion was performed with NgoMiV and AgeI, producing complementary overhangs. Religated pSB1C3 (due to incomplete vector dephosphorylation) loses the AgeI recognition sequence. Therefore plasmids without insert should run in the supercoiled position at about 1.5 kb, plasmids with inserts should run in the linearised position at 3 kb. Single enzyme digestion was chosen, because the insert is too small to be solved on a gel.
Procedure:
- analytic digestion: 3 µl of plasmid DNA P32 to P35 (about 300 ng) were digested with 0.3 µl AgeI-HF in 1 µl CutSmart Buffer and 5.7 µl H2O. The reaction was incubated for 2 h at 37°C.
- 5 µl digestion mix were loaded on a 1% agarose gel for electrophoresis, 1 kb ladder
Result:
- P32 to P35 might carry the insert because they all appear in the linearized position at 3 kb
- Therefore plasmid sequencing should confirm the result.
Lane 1: 1 kb ladder Lane 3: P32 Lane 4: P33 Lane 5: P34 Lane 6: P35
Week 3 (May 30th - June 5th)
Monday, May 30th
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CR
Aim of the experiment: Sequencing of P32 and P35
Procedure:
- Sequencing batches were prepared according to manufacturers protocol (Mix2Seq Kit, Eurofins Genomics): 15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer (10 µM)
- The plasmids received the following barcodes:
- P32 (VF2) : FR11326637
- P35 (VF2) : FR11326636
Religation of F13 (digest of pSB1C3 with AgeI and NgoMIV) and transformation in XL1 blue
Investigator: CR
Aim of the experiment: Re-ligate digested and dephosphorylated pSB1C3 and transformation in XL1blue
Procedure:
- Plasmid was ligated with the T4 DNA Ligase following the manufacturer’s protocol (NEB)
- 52 ng Vector DNA were used
- The Ligation mix was incubated for 30 min at RT
- 7 μL Ligation mixed were used for the transformation of 50 μL XL1 blue.
- Transformation was performed following the SOP
Result:
- 5 clones on plate
Inoculation of pre-cultures with B0034 (strong RBS), K577894 (TET repressed YFP), KK577895 (Tet repressed RFP), F13+F14
Investigator: CR
Procedure:
- Add 50 µL ampicillin(B0034)/chloramphenicol(K5777894, K577895, F13+F14) to 50 mL LB-medium
- Picking colonies from plate (XL1 blue; all rescue)
- Incubate over night at 37°C
Tuesday, May 31th
Dialysis of eGFP
Investigator: NA
Aim of the experiment: Changing the buffer of eGFP for biotinylation (from TRIS to PBS)
Procedure:
- eGFP was poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in ice cold PBS pH 7.4
- The dialysis took place at 4°C overnight
Expression and harvest of Streptavidin (pASK75 (SA1)) in BL21 in TB-medium
Investigator: JB, NA
Aim of the experiment: Recombinant expression and purification of Streptavidin
Procedure:
- The pellet was transferred into a beaker of sufficient size and resuspended in fridge-cooled Tris Buffer B (50 mM Tris/HCl (pH = 8.0), 1 mM EDTA)
- The solution was homogenized in the PANDA (see SOP)
- The resulting lysate was transferred into centrifuge tubes and spun down (4°C, 18,000 rpm, 30 mins, SS34-rotor). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight.
Transformation of P30 (Tet repressed GFP)
Investigator: NA
Aim of the experiment: expression of tet-repressed GFP in E. Coli BL21
Procedure: transformation in competent E. Coli BL21 according to protocol
Result: plates (LB Cam) in incubator for further processing (37 °C)
Cloning of A3C5 and Avi-Tag into pSB1C3
Investigator: CR, JH
Aim of the experiment: Preculture of pSB1C3 (F13+F14) was red See if RFP-generator is still inside the pSB1C3 See if ligation of EspP was successful
Procedure:
- MiniPrep of P39 was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- pSB1C3 was digested with AgeI and NgOMIV (1.5 µL P39 + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 22.5 µL ddH2O; incubation for 2h, 37°C)
- FastAP was added and the reaction was incubated for another 10min at 37°C
- FastAP was inactivated by heating the mix to 75°C for 10min.
- The digestion was purified by gelelctrophoresis (1%, 75V, 1h)
Results:
Unknown signals. Repeat transformation, digestion, dephosphorylation and ligation of pSB1C3
Re-transformation of P4 (pSB1C3 (RFP-generator)) in XL1 blue
Investigator: JH, CR
Aim of the experiment: Transformation
Procedure:
- 2 μL P4 were used for transformation of XL1 blue
- Transformation was performed according to SOP (incl. rescue plate)
Competent E. coli XL1 cells
Investigator: JB
Aim of the experiment: Generation of competent E.coli XL1 cells
Procedure:
- 5 ml LB w.o. antibiotics (sterile)
- Inoculation of one colony of E.coli XL1 cells
- Incubate overnight at 37°C
Wednesday, June 1th
eGFP Biotinylation
Investigator: CG
Aim of the experiment: Biotinylation of dialysed eGFP past IEC
Procedure:
- The average concentration of the pooled factions 12 and 13 should be about 0.5 mg/ml (=19 µM)
- 10 ml of a 100 mM Biotin stock-solution was prepared (340 mg)
- 3.5 ml of eGFP were biotinylated with 12 µl 100 µM Biotin-solution (20x molar excess)
- Incubation at 4 °C overnight
pSB1C3 vector backbone preperation
Investigator: CG
Aim of the experiment: Inoculation of two colonies pSB1C3-RFP Generator
Procedure:
- 2x 5 ml LB+Cam medium (1:1000)
- Picking colonies from plate (XL1 blue, rescue)
- Incubate overnight at 37°C
Generation of GFP expressing E. coli cells
Investigator: CG
Aim of the experiment: Inoculation of BL21 transformed cell with P30
Procedure:
- 50 ml LB+Cam medium (1:1000)
- Picking colonies from plate (BL21, rescue)
- Incubate over night at 37°C
Competent E. coli XL1 cells
Investigator: JB
Aim of the experiment: Generation of competent E. coli XL1 cells
Procedure:
- Inoculated 500 µl preculture in 50 ml LB media w.o. antibiotics
- Incubated and shaken at 37°C until the OD550 reached 0.44
- Transferred into a falcon tube and incubated on ice for 10 mins
- Spun down at 3000 g for 10 mins at 4°C, supernatant cast away
- Pellet resuspended in 40 ml fridge-cooled MgCl2 (100 mM), once again spun down at 4°C for 10 mins at 3000 g
- Supernatant cast away, pellet resuspended in 20 ml cold CaCl2 (50 mM)
- Incubation on ice for 30 mins
- Centrifugation repeated, supernatant cast away
- Pellet resuspended in 2 ml of CaCl2 (50 mM) and glycerol (15%)-solution, aliquoted (50 µl each), frozen in liquid nitrogen and stored at -80°C for further use
Cloning of Pcat-RBS-construct in pSB1A2
Investigator: CG, JH, CR
Aim of the experiment: Pcat (I14033) was cut out of P27 and ligated into P38 for generating the Pcat-RBS (B0034) construct.
Procedure:
- P27 was digested with EcoRI and SpeI (4 µL P27 + 1.5 µL EcoRI + 1.5 µL SpeI + 3 µL CutSmart + 20 µL ddH2O; incubation for 2h, 37°C)
- P38 was digested with EcoRI and XbaI (10 µL P27 + 1.5 µL EcoRI + 1.5 µL XbaI + 3 µL CutSmart + 14 µL ddH2O; incubation for 2h, 37°C)
- Preperative gel electrophoresis was performed
- For P27 the signal at about 50 bp was cut out and gel extracted according to manufacturer's protocol (Gelextractionkit, Qiagen): 5.9 ng/µl
- For P30 the signal at about 2.1 kb was cut out and gel extracted according to manufacturer's protocol (Gelextractionkit, Qiagen): 8.9 ng/µl
- Ligation according to SOP
Purification of streptavidin
Investigator: JB
Aim of the experiment: Refolding denaturized streptavidin
Procedure:
- the GdmCl-solved streptavidin was spun down using an SS-34 rotor (~60 min, 18,000 rpm, 4°C)
- The supernatant containing the solved and denaturized protein was transferred into a falcon tube, the pellet cast away
- The protein solution was slowly dripped into 4 l of cooled 1x PBS (in the 4°C room) using a hydraulic pump and stirred overnight
Tuesday, June 2th
Preparation of pSB1C3 for further use
Investigator: CR
Aim of the experiment: Digestion of pSB1C3 to remove RFP-generator
Procedure:
- MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- P40 was digested with AgeI and NgOMIV (2.5 µL P40 + 1.5 µL AgeI + 1.5 µL NgoMIV + 3 µL CutSmart + 21.5 µL ddH2O)
- Incubation over night at 37°C
Generation of GFP expressing E. coli cells
Investigator: CG, JH
Aim of the experiment: Various inductor concentrations
Procedure:
- 12x 5 ml LB+Cam media were inoculated with 50 µl preculture
- when cultures reached OD550 of 0.5, they were induced with tetrazycline-anhydride of various concentrations (0 ng/µl, 20 ng/µl, 40 ng/µl etc., 200 ng/µl)
- t=0, =30, =90, =150, =210, =270 a 100 µl aliquot of each culture was stored at 4 °C in the refrigerator
Dialysis of biotinylated eGFP
Investigator: JH, JB
Aim of the experiment: Purification of biotinylated eGFP
Procedure:
- eGFP was poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in 2 L ice cold Tris/HCl 20 mM pH 8.0
- The dialysis took place at 4°C over night
Ligation of F17.1 (p(cat)) and F18 (stron RBS) and transformation in XL1 blue
Investigator: JH
Aim of the experiment: Ligation and transformation in XL1blue
Procedure:
- Plasmid was ligated with the T4 DNA Ligase following the manufacturer’s protocol (NEB)
- 9 µL F18 (B0034 in pSB1A2; RBS) and 1 µL of F17.1 (I14033; P(cat)) were used [AmpR]
- The Ligation mix was incubated for at least 2h at RT
- 7 μL Ligation mixed were used for the transformation of 50 μL XL1 blue
- Transformation was performed following the SOP [AmpR], incl. rescue plate
Friday, June 3th
Cloning of F8 (A3C5), F9 (Avi-Tag) and F14 (EspP) into F20
Investigator: CR
Aim of the experiment: Cloning of A3C5, Avi-Tag and into pSB1C3
Procedure:
- FastAP was added and the digestion of F20 and was incubated for another 15 min at 37°C
- The digestion was purified by gelelctrophoresis (1%, 75V, 1h)
- Plasmid backbone (signal 1) was cut from the gel
- Gelextraction was performed after manufacturer's protocol (Gelextractionkit, Qiagen)
Results:
Signal 1: F20
[F20]= 13,8 ng/μL
Generation of GFP expressing E. coli cells
Investigator: CG, NA
Aim of the experiment: SDS PAGE for verification of successful GFP expression
Procedure:
- SDS electophoresis according to SOP
- Lane 1: #12, 90 min
- Lane 2: #12, 150 min
- Lane 3: #12, 210 min
- Lane 4: #12, 270 min
- Lane 5: 8 µl
Results:
Inoculation of colonies from F19 (pcat-strong RBS)
Investigator: CG
Aim of the experiment: three colonies were picked from yesterdays transformation
Procedure:
- 3x 5 ml LB+Amp media
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
Cloning of F8 (A3C5), F9 (Avi-Tag) and F14 (EspP) into F20 (pSB1C3)
Investigator: CG
Aim of the experiment: Ligation of F21, F22 and F23 into F20 and transformation
Procedure:
- Ligation was performed according to the SOP with: 0.7 µl F8 (1:10) / 9.3 µl F20, 0.7 µl F9 (1:10) / 9.3 µl F20, 9.1 µl F14 / 0.9 µl F20
- Transformation was performed according to the SOP on LB Cam-Agar
Precipitation of Streptavidin
Investigator: NA, JB, LK
Aim of the Experiment: Purification of Streptavidin
Procedure:
- Slowly add (NH4+)2SO42- until the concentration reaches 40% (23,1 g/100mL) while stirring
- Let it stir over night to ensure complete precipitation
- Centrifuge at 8500 rpm at 4°C for 15min (rotor: SLA1500), dismiss the pellet and use the supernatant for further processing
- Slowly add (NH4+)2SO42- until the concentration reaches 70% (19,1 g/100mL) while stirring
- Let it stir over night (slowly to avoid foaming)
- Centrifuge at 8500 rpm at 4°C for 15min (rotor: SLA1500), dismiss the supernatant and use the pellet for further processing
- Resuspend the protein pellet in a minimum volume of TRIS/HCl; pH 8; 20mM (NO SALT) (6 mL per pellet), the pellets where then united and titrated with Buffer until the solution became clear. Total amount of buffer: 40mL
- Transfer to 50 mL Falcon Tubes and centrifuge at full speed for 15 min. Filter the supernatant through 45nm sterile filter and keep the pellet (rescue).
Saturday, June 4th
Inoculation of colonies from F21 (pSB1C3 (A3C5)), F22 (pSB1C3 (Avi-Tag)), F23 (pSB1C3 (EspP))
Investigator: NA
Aim of the experiment: three colonies were picked from each transformation
Procedure:
- 3x 5 ml LB+Cam media for each fragment
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
MiniPrep of F19 (pSB1A2 (pcat-strong RBS))
Investigator: NA
Aim of the experiment: Extraction of Ligation: F17+F18 (starke RBS+P(cat))
Procedure: MiniPrep was performed after manufacturer's protocol (QIAprep MiniPrep, Qiagen) Concentrations: K1= 66,2 ng/µl; K2= 60,2 ng/µl; K3= 104,4 ng/µl
Week 4 (June 6th - June 12th)
Monday, June 6th
Cloning of TetR from p28 into p31 (pSB1C3(weak RBS))
Investigator: LK, JH, NA
Aim of the experiment: Cloning TetR in front of weak RBS in pSB1C3
Procedure:
- Digestion of p28 with EcoR1 and Spe1 (20µl DNA, 2,5 µl each enzyme, 5µl CutSmart buffer, 20µl ddH2O) and p31 (4µl DNA, 1µl EcoRI / XbaI, 2µl, 12µl DNA) -> Incubation at 37°C for 2h.
- Electrophoresis: 1% Agar, 15 min , 70 V (oben p31 und p41, unten p28)
- Gelextraction concentrations: p28 (F25) = 2,3 ng/µl
p31 (F24) = 7,8 ng/µl
- Ligation with 6,3 µl vector (F24) and 1,7 µl Insert (F25) + 1µl Buffer + 1µl Ligase over night, 16°C
SDS-PAGE of Streptavidin
Investigator: NA, JH, LK
Procedure: mixing of 80 µl sample with 20 µl SDS buffer and heating at 95°C for 10 min. 1 h staining, 1 night unstaining. Concentration of streptavidin was measured via UV/VIS spectroscopy.
c = 23,5 mg/ml
Result:
- 1. Lane: 8 µl Marker
- 2. Lane: 5 µl of Streptavidin_1:10
- 3. Lane: 5 µl of Streptavidin_1:50
- 4. Lane: 5 µl of Pellet_1:500
- 5. Lane: 5 µl of after 40% precipitation_1:10
- 6. Lane: 5 µl of supernatant after 70% precipitation and centrifugation
Successful Streptavidin production verified.
Sequencing of P41 (pSB1C3 (strongRBS-p(cat)))
Investigator: NA
Aim of the experiment: Sequencing of P41
Procedure: Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (100 ng/ µl) and 2 µL sequencing primer (VF2))
The plasmids prepared received the following barcode:
- P41 : FR11326633
Tuesday, June 7th
Transformation of P30 (Tet repressed GFP), P36 (Tet repressed RFP), P37 (Tet repressed YFP)
Investigator: NA
Aim of the experiment: Transformation of RFP, GFP and YFP in BL21
Procedure: transformation in competent E. Coli BL21 according to protocol + rescue
Result: plates (LB Cam) in incubator for further processing (37 °C)
MiniPrep of P42 to P50, analytic digestion, sequencing
Investigator: CG
Aim of the experiment: Verification of successful cloning
Procedure: MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Concentrations (ng/µl each) for P42: 260, P43: 247, P44: 181, P45: 118, P46: 261, P47: 135, P48: 148, P49: 240, P50: 299
- Analytic digestion: 0.3 µl AgeI, 2 µl CutSmart, 6.7 µl H2O for P42, P43, P44, P45, P46, P49, P50 and 0.3 µl EcoRI, 0.3 µl PstI, 2 µl CutSmart, 6.4 µl H2O for 2 h at 37°C
- 1% agarose gel electrophoresis
- Sequencing: P42= FR11326632 (VF2), P45 =FR11326631 (VF2)
Results:
- Lane 1: 5 µl 1 kb Ladder
- Lane 2-10: Digestion of P42 to P50. P47 and P48 are carrying the insert. All other plasmids are religated and lost their AgeI site, hence they run in the ccc position.
Dialysis of not biotinylated eGFP
Investigator: NA
Aim of the experiment: Purification of eGFP for mass determination
Procedure:
- eGFP was poured in a dialysis tubes
- The tubes were then placed in ice cold ammonium acetate
- The dialysis took place at 4°C over night
Polymerisation attempts
Investigator: JH, CR, CG, JB
Aim of the experiment: Identification of optimal polymerisation conditions for biotinylated eGFP and BSA in a Streptavidin reservoir
Procedure:
- variate of compositions for both solutions was examined to modify density properties during “printing”
- all compositions can be found in excel sheet
Results:
- no or little successful polymerization at Streptavidin concentrations of 4 mg/mL and below; immense density incompatibilities
- sucessfull polymerization at Streptavidin concentrations of 10 mg/mL and BSA solution with approx. 15% glycerol
- no success with biotinylated eGFP (too few Biotin molecules?!)
Wednesday, June 8th
Sequencing of P36 (Tet repressed RFP) and P37 (Tet repressed YFP)
Investigator: NA
Aim of the experiment: Sequencing of P36 and P37 in both directions
Procedure: Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- P36 (VF2): FR11326630
- P36 (VR): FR11326629
- P37 (VF2): FR11326628
- P37 (VR): FR11326627
Dialysis of Streptavidin
Investigator: NA, JH
Aim of the experiment: Purification of Streptavidin (possible ammonium sulfate contamination after precipitation)
Procedure:
- Streptavidin was poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in 3 L ice cold Tris/HCl 20 mM pH 8.0 (no salt)
- The dialysis took place at 4°C over night
Polymerisation attempts
Investigator: JH, JB
Aim of the experiment: Identification of optimal polymerisation conditions for biotinylated BSA in a Streptavidin reservoir
Procedure:
- variate of compositions for both solutions was examined to modify density properties during “printing” (Streptavidin at 10 mg/mL or higher)
- all compositions can be found in excel sheet
Thursday, June 9th
Generation of GFP, YFP, RFP expressing E. coli cells
Investigator: CG
Aim of the experiment: Various inductor concentrations, various fluorescent proteins were expressed in E coli cells
Procedure:
- 3x 5 ml LB+Cam media were inoculated with 50 µl preculture of GFP (P30), RFP (P36), YFP (P37)
- when cultures reached OD550 of 0.5, they were induced with anhydro-tetrazycline of various concentrations (0 ng/µl, 100 ng/ml, 200 ng/ml)
- 50 ng/ml = 12,5 µl of 1:100 2 mg/ml stock solution, 200 ng/ml = 50 µl of 1:100 2 mg/ml stock solution
- t=0, =4 h and stored on ice
Resuts: no fluorescent could be detected. Therefore an overnight induction at 37°C with 1 µM/ml inductor concentration was performed with the RFP plasmid.
Biotinylation of GFP expressing E. coli cells
Investigator: CG
Aim of the experiment: Biotinylation of P30 carrying E. coli cells
Procedure:
- 850 µl P30 preculture from Thursday 2nd were centrifuged and the supernatend was discarded
- Pellet was resuspended in 800 µl PBS-buffer
- 100 µl 100 mM Biotin-NHS was added
- incubation for 4 h at room temperature
- Experimenting with some glycerol and streptavidin concentrations. A more structured approach will be done the next day.
Results: A shape thin “sausage” could be detected. Therefore a bigger E. coli culture was biotinylated at 4°C overnight for experimenting with the next day.
Cloning of TetR promotor and weak RBS in pSB1C3
Investigator: CR, JB
Aim of the experiment: MiniPrep and NanoDrop of F26 K1, K2 and K3 Sequencing of P51
Procedure: MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen) Concentrations were as following:
- P51: 163,3
- P52: 151,7
- P53: 140,5
P51 was sequenced:
- 1. with VF2: FR11326626
- 2. with VR: FR11326625
Biotinylation of eGFP
Investigator: VL, JB
Aim of the experiment: Biotinylation of eGFP with 200x biotin-NHS molar excess over eGFP (15x excess over surficial K residues). NB: previous trial unsuccessful. Excess of biotin was likely insufficient to ensure efficient biotinylation, esp. since biotin-NHS slowly hydrolyzes in the aqueous environment.
Procedure: 1 ml of eGFP was biotinylated with an 200x excess of biotin in DMF (100 uM; 38 uL), 5.5 h at rt. The solution was dialysed versus 20 mM Tris/HCl pH 8 overnight at 4 °C.
PCR of gene syntheses
Investigator: LK
Aim of the experiment: Amplification od iGEM_1_SA-m1 (F27), iGEM_1_mRuby_EGFR (F28) and iGEM_2_IRES-BirA (F29)
Procedure:
- Add 100µl of ddH2O to the gene sythesis samples and incubate for 20 min at 50°C
- Dilute samples to a final concentration of max. 1ng/µl (1:10) with water.
- Approach:
1µl DNA (diluted)
5µl buffer
0,5µl dNTPs
1,25µl forward Primer VF2
1,25µl reverse Primer VR
0,25µl Q5 Polymerase
Fill up with water up to 25µl
- Make sure to pipette everything on ice and add the polymerase at the end
- Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/)
98°C_1min initiation step
98°C_10sec
66°C_30sec > step 2-4: 35 repeats
72°C_30sec
72°C_2min final step
4°_hold
- The samples were then prepared with the PCR Product Purification Kit
Polymerisation experiments for biotinylated eGFP and BSA with Streptavidin
Investigator: JH, CR, JB
Aim of the experiment: Determination of ideal concentrations for both protein interaction partners as well as buffer viscosity (glycerol content etc.) for polymerisation in a test tube
Procedure:
- Different concentrations of proteins and glycerol were tested to support stable polymerisation without protein aggregates rising to the surface or sinking to quickly
- The experiments done are not going to be further documented as the streptavidin used for the experiments was previously determined in a wrong fashion and was actually considerably lower than expected (4x lower). Streptavidin was, due to that circumstance, concentrated to be able to support experiments with at least 5 mg/ml concentrations
- First experiments with concentrated streptavidin seemed promising, though a complete series of experiments with varying parameters should be conducted in the future
Sequencing of P29 (pSB1C3 (double terminator), P36 (pSB1C3 (Tet repressed RFP)) and P37 (pSB1C3 (Tet repressed RFP))
Investigator: JB
Aim of the experiment: Sequencing of P29, P36 and P37
Procedure: According to the manufacturer's protocol.
- P29: FR11326624 (VF2)
- P36: FR11326623 (VF2)
- P37: FR11326621 (VR), FR11326622 (VF2)
Inoculation of (pre)cultures for a pSB1C3 midiprep
Investigator: JB, VM
Aim of the experiment: Massively amplifying the pSB1C3 vector (which will be needed for building up new biobricks) by midiprepping the P40 plasmid (RFP-generator in pSB1C3). Similar to minipreps, but on a greater scale. Procedure: 3 ml precultures were inoculated with a clone from the plate previously used for minipreps. After the precultures had grown for 4 hours (due to shortage of time), 2 x 50 ml cultures were inoculated and shaken overnight at 37°C. The next day, cultures were put into the fridge for storage until Monday.
Week 5 (June 13th - June 19th)
Monday, June 13th
Digestion of gene synthesis iGEM1 and iGEM2
Investigator: LK, JH
Aim of the experiment: Digest F28 (iGEM1) and F29 (iGEM2) in order to ligate them into P40, which was equally digested (to get rid of the rfp-generator and have pSB1C3 as backbone)
Procedure:
- 30µl of F28 were digested for 3h with SpeI and NgoMIV, and 30 µl of F29 were digested with XbaI and PstI
- 2 x 20 µl where digested with the equivalent enzymes for 3h
- After electrophoresis the bands were cut out and purified
Results: Results show that PCR of F28, F29 might not have worked. P40 was extracted from gel (F30 and F31). PCR of gene synthesis was repeated (50µl!)
Midiprep of P40
Investigator: LK
Aim of the experiment: Purification of P40 from
Procedure:
- See protocol of Midiprep kit
- 1xuse-centrifugation-columns broke in centrifuge during last step, DNA rescue was tried by resuspending rest of the solution in the tube in EB buffer and redo the last steps (isoprop and ethanol precipitation)
- Airdry "pellet"(no pellet visible) for 15 min under fume hood and resuspend in 100µl EB Buffer
- Measure concentration with nanodrop
Preparation of Collagen + Biotinylation
Investigator: VL, JL
Aim of the experiment: prepare collagen solution + biotinylate small sample
Procedure:
- Dissolve 5 g collagen in 50 mL Tris/HCl pH 8 20 mM w/o salt --> 770 uM solution (100 mg/mL)
- Sterile filter
- Biotinylation: 980 uL Tris buffer + 20 uL collagen solution + 30 uL biotin-NHS (100 mM in DMF, 200x molar excess over collagen monomers)
- Reaction time:
- Dialyze 4 °C Tris/HCl pH 8 20 mM w/o salt o/n
Transformation of P9 (pASK75)
Investigator: JL, VL
Aim of the experiment: transform streptavidin expression vector P9 (clone 4) into E. coli BL21
Procedure:
- Standard (see booklet): 45 s at 42 °C; incubate 1h, 37°C; plate: medium w. chloramphenicol (+ rescue plate) Incubate o/n 37°C.
- Wrong antibiotic plate, repeated the next day of the experiment: Massively amplifying the pSB1C3 vector (which will be needed for building up new biobricks) by midiprepping the P40 plasmid (RFP-generator in pSB1C3). Similar to minipreps, but on a greater scale.
Tuesday, June 14th
Mass spectrometry of biotinylated eGFP
Investigator: CR
Aim of the experiment: identify biotinylation degree of eGFP
Procedure:
[generic_display_report] (i could not upload this file, sorry.)
Results: about 4-5 biotins per protein. To verify the biotinylation degree the unbiotinylated eGFP will be measured.
Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3
Investigator: CR
Aim of the experiment: Purification of PCR products
Procedure:
- Volume of PCR reaction: 50 µL
- PCR products were purified by manufacturers protocol (QIAquick PCR purification kit, Qiagen)
- Concentration measured by Nanodrop:
- F32: 87,5 ng/µL
- F33: 68,3 ng/µL
- Test if PCR worked:
- Samples were applied on a 2% agarose gel and was run at 80 V for 40 min.
Results:
- PCR of F28 worked --> F32
- PCR of F29 did not work --> wrong primers were used for amplification --> F33 not right (Re-Do PCR with appropriate primers
Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3
Investigator: CG
Aim of the experiment: Digestion of F32 (SpeI, NgoMIV) and F33 (XbaI, PstI)
Procedure:
- Digestion F32: 27 µl of F32 (2.4 µg), 5 µl CutSmart Buffer, 1.5 µl SpeI, 2 µl NgoMIV, 14.5 µl H2O
- Digestion F33: 27 µl of F32 (1.8 µg), 5 µl CutSmart Buffer, 1.5 µl XbaI, 1.5 µl PstI, 15 µl H2O
- Incubation for 3 h at 37°C
- Preperative gel:
- Gelextraction of F32 upper band and F32 lower band was executed by manufacturers protocol (Qiagen)
- F32 upper band (mRuby) was named F34 (=10 g/yl)
- F32 lower band (eGFR) was named F35 (=6 ng/yl)
Cloning of F32 (PCR iGEM_1 mRuby-EGFR) and F33 (PCR iGEM_2 IRES-BirA) in pSB1C3
Investigator: CR
Aim of the experiment: Ligation of F34 (mRuby) and F35 (eGFR) in F30 (pSB1C3); Tranformation in XL1blue
Procedure:
- Ligation mixtures were prepared as following:
- F34: 8.0 μL F34, 2 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase)
- F35: 7.6 μL F35, 2.4 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase)
- Ligation was incubated for 30 min at RT
[something is missing here]
Re-Sequencing of P37 (Tet repressed YFP)
Investigator: CG
Aim of the experiment:
Purification and sequencing of P30
Procedure:
- P30 was heated up to 100 °C and centrifugated afterwards to remove proteins from Promotor (if there are some), the supernatant is sequenced
- Sequencing batches were prepared after manufacturer's protocol
- (15 μL plasmid DNA (50-100 μM) and 2 μL sequencing primer)
- The plasmid we prepared received the following barcode: P37 (VF2) : FR11326620
Streptavidin expression
Investigator: CR
Aim of the experiment: Transformation of P9 (pASK74 (SA1)) in BL21
Procedure:
- Transformation was executed following SOP
- Transformed cells were plated on LB medium (Amp) (50 µL and rescue (50 µL)
Dialysis of Streptavidin and biotinylated BSA
Investigator: CR
Aim of the experiment: Streptavidin and bitinylated BSA are both in Tris/HCl without salt. SInce we will perform later experiments in salty solutions the dialysis was performed against 137 mM NaCl (Saltconcetration of PBS)
Procedure:
- 3 L Tris/HCl NaCl solution was prepared
- Dialysis was prepared according to SOP
- Cut off Dialysis hose: 14 kDa
Inoculation of (pre)cultures of P40 for a pSB1C3 midiprep [repeat]
Investigator: JH
Aim of the experiment: Massively amplifying the pSB1C3 vector by midiprepping the P40 plasmid (RFP-generator in pSB1C3)
Procedure: four 4 ml precultures were inoculated with a clone from the plate previously used for minipreps. After the precultures had grown for 6 hours, 50 ml cultures were inoculated and shaken overnight at 37°C.
Subcloning of P41 (strong RBS-p(cat)) Insert into pSB1C3
Investigator: JH
Aim of the experiment: Moving P41 (pSB1A2; Pcat+weakRBS) to pSB1C3 for biobrick submission
Procedure:
- Digestion of P41 with XbaI and PstI (10 µl DNA, 1,5 µl each enzyme, 3 µl CutSmart buffer x10, 14 µl ddH2O) -> Incubation at 37°C for 2h.
- Electrophoresis: 2% Agar, 40 min , 90 V (1kb ladder, empty, P41, 100b ladder)
- GelExtraction
Resuts: no visible signal of insert at approx. 100b. Area was cut blind for GelEx. No DNA in the sample according to NanoDrop measurement. Repeated the next day.
Inoculation of colonies from NanoLuc
Investigator: JH
Procedure:
- 2x 4 ml LB+Cam media
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
Wednesday, June 15th
Subcloning of P41 (strong RBS-p(cat)) Insert into pSB1C3
Investigator: CG
Aim of the experiment: Moving P41 (pSB1A2; Pcat+weakRBS) to pSB1C3 for biobrick submission. No gel extraction was performed because the fragement is too small to visualize on the gel.
Procedure:
- Digestion of P41 with XbaI and PstI (10 µl DNA, 1,5 µl each enzyme, 3 µl CutSmart buffer x10, 1 µl FastAP, 13 µl ddH2O) -> Incubation at 37°C for 2 h, heat inactivation at 80°C for 20 min.
fragement is to small to visualize on agel.
- Subcloning into F31: 3.8 μL F38, 6.2 μL F31, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase according to the SOP
- Transformation into E. coli XL1 according to the SOP
Miniprep of pSB1C3(Nanoluc)
Investigator: LK
Aim of the experiment: Preperation of a BioBrick with NanoLuciferase in pSB1C3
Procedure:
- The Miniprep was done according to the protocol.
- The two tubes where pooled.
- The new Plasmid is found under P54 and was sent to sequencing.
Polymerisation
Investigator: EU
Aim of the experiment: Determine optimal polymerisation conditions for BSA and Collagen
Procedure: The experiment was performed in a 96 Well plate:
Results: maximum concentration of Streptavidin and biotinylated Protein works best. Collagen does not work. Glycerolconcentrations do not inhibit interaction. Reaction might be to slow at given concentrations to be effective in printing process.
MidiPrep of pSB1C3 with RFP generator (P4)
Investigator: NA
Aim of the experiment: purification of pSB1C3 with RFP generator for further processing (P56)
Procedure:
- MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen)
Results: Concentration very low (12ng/μL), but high volume (500 μL)
Thursday, June 16th
MiniPrep of F36 (Ligation mRuby in pSB1C3) K1 & K2, F37 (Ligation EGFR in pSB1C3), I13453 (pBad promotor) K1 & K2, K577881 (pC+AraC+pBad+GFPc) K1 & K2
Investigator: CR
Aim of the experiment: isolate Plasmids
Procedure:
- MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Plasmids were named as following: [concentration]
F36 K1: P57 [212,9 ng/μL]
F36 K2: P58 [181,3 ng/μL]
F37: P59 [195,5 ng/μL]
K577881 K1: P60 [557,2 ng/μL]
K577881 K2: P61 [791,9 ng/μL]
I13453 K1: P62 [130 ng/μL]
I13453 K2: P63 [460 ng/μL]
Sequencing of P29 (double terminator), P36 (Tet repressed RFP) and P37 (Tet repressed YFP)
Investigator: JB
Aim of the experiment: Sequencing of P56, P57, P59, P61 and P62
Procedure: According to the manufacturer's protocol.
- P56: FR11326618 (VF2)
- P57: FR11326617 (VF2)
- P59: FR11326616 (VF2)
- P61: FR11326613 (VR), FR11326615 (VF2)
- P63: FR11326614 (VF2)
MidiPrep P4 (pSB1C3)
Investigator: VL
Aim of the experiment: isolate Plasmids
Procedure:
- MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen). Centrifugation steps at 20,000 g performed at 12,500 rpm in special centrifugation tubes. Since only one column was left, both prepped cell pellets were applied to one column. Flow-through and wash was collected.
- Conc:
Amplification of BioBricks (overnight cultures for minipreps)
Investigator: JB
Aim of the experiment: Inoculation of 5 ml overnight cultures for minipreps. The following biobricks were used:
- BBa_K157012
- BBa_K243004
- BBa_K243006
- BBa_K157001
- BBa_K243005
- BBa_I712009
- BBa_K404108
- BBa_K782061
- BBa_K1371012
- BBa_K782060
Procedure:
- Three hours of incubation of the tubes containing the bacterial agar at 37°C (Note: 12 hours are actually recommende)
- Inoculation of two times 5 ml preculture containing the appropriate antibiotic per clone. For KanRAmpR (reistance against both ampicillin and kanamycin) it was only selected for ampicillin.
- Incubation shaking overnight at 37°C
PCR Amplification of genesynthesis 2 (F29; IRES-BirA)
Investigator: LK
Aim of the experiment: Amplification of Genesynthesis_2 (IHRES_BirA_StrepTag)
Procedure: A 50µl approach was made:
- 33,5 µl Water
- 2µl DNA (F29, diluted 1:10)
- 2,5µl of each Primer (O11 and O12 c:10µM)
- 0,5µl dNTPmix
- 5µl Reaction buffer
- 0,5µl Q5-Polymerase
- The Annealing Temp. was set an 58°C
- Next step is to run an analytic gel (band should be visible at 1,5 kB)
Friday, June 17th
Cloning of genesynthesis 2 (IRES-BirA) into pSB1C3
Investigator: CG, NA, JH
Aim of the experiment: Purification oft he PCR reaction, gel verification, restriction digestion (F29), ligation with F31 and transformation
Procedure:
- Yesterdays PCR was purified according to Qiagen PCR purification kit
- An anlaytic gel electrophoresis was performed on a 1% gel with 4 µl purified PCR product
- Restriction digest: 26 µl purified PCR product, 1.5 µl XbaI, 1.5 µl PstI, 5 µl NEB SmartCut, 16 µl H2O. Incubation for 2 h at 37°C.
- Gel extraction
- Ligation: F31
- Transformation
Results: Gel electrophoresis after PCR purification exhibit a thin signal at 1.5 kb. Next time 1:30 min for elongation time might increase the signal.
Miniprep new iGEM BioBricks
Investigator: VL
Aim of the experiment: Purify plasmid DNA from transformed cells
Procedure:
- Miniprep according to manual (pellet centrifuged 5000 rpm, 10 min, 4 °C)
- Elution with 50 uL EB Buffer
- Concentrations (all ng/uL):
K782061 (a) 172.9, (b) 53.0
K243006 (a) 241.1, (b) 58.6
K1371012 (a) 138.0, (b) 43.0
K157001 (a) 184.3, (b) 186.4
K782060 (a) 339.0, (b) 254.0
K243005 (a) 62.3, (b) 176.2
K243004 (a) 122.3, (b) 110.3
I711009 (a) 185.2, (b) 174.7
K157012 (a) 181.2, (b) 195.3
K404108 (a) 116.4, (b) 142.5
MidiPrep of pSB1C3 with RFP generator (P4)
Investigator: NA
Aim of the experiment: purification of pSB1C3 with RFP-Reportercasette for further processing (P74)
Procedure: MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen)
Results:
Cloning of IRES-BirA (F33->F40) into digested F31 (pSB1C3)
Investigator: JH, LK
Aim of the experiment: Ligation of IRES-BirA (F29->F33->F40) into F31 (digested P40 by XbaI and PstI = pSB1C3 without insert), gelelectrophoresis and tranformation in XL1blue
Procedure:
- F33 (PCR product of F29/gen synthesis 2) was digested with XbaI and PstI
- Gelextraction of band at 1500 bp was executed by manufacturers protocol (Qiagen)
- intermediate result: F40 with 6.6 ng/µL
- Ligation mixtures were prepared as following:
- 9.6 µL F40, 0.4 µL F31, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- Transformation: (Saturday)
- Transformation of Ligation (F29+F31) into XL-1 blue was done according to the SOP. The plate was incubated until Sunday at 37°C.
- 3mL-cultures in LB+ 30µg/ml cam where inoculated on Sunday
- Mini-prep according to manufacturer's protocol was done on Monday.
PCR, purification and digestion of P19 (EspP) and F28 (mRuby EGFR) for BioBrick creation
Investigator: JB, LK
Aim of the experiment: Amplification of EspP from P19 and mRuby/EGFR (F28) for creation of Biobricks
Procedure (for both fragments):
- PCR according to the SOP (primers for P19: O13 and O03, primers for F28: VR and VF2)
- PCR purification via the purification kit according to the manufacturer's protocol (concentrations: P19(EspP):119,5 ng/µl F28(mRuby):56,2ng/µl
- Analysis on an 1% agarose gel using 5 µl purified PCR product
- Restriction digestion with NgoMIV/SpeI (for both cases)
- 40µl DNA
- 1,5µl of each Enzyme
- 2µl ddH2O
- 5µl CutSmart Buffer
- The tubes where incubated at 37°C for ~2 days.
Results: Analytical gel electrophoresis after PCR purification shows thick bands at roughly 1000 bp
Still to do:
- Gel extraction of both signals for F28 and one signal for EspP-PCR
- Ligation with appropriate vector (e.g. F30)
- Transformation
Minipreps of F39 (strong RBS-pcat in pSB1C3)
Investigator: JB
Aim of the experiment: Amplification of F39 (ligation, pCat with strong RBS)
Procedure:
- Miniprep according to the manufacturer's protocol
- Resulting plasmid: P76
- Concentration: 52 ng/µl
Minipreps of pSB1C3 with RFP generator (P4)
Investigator: JH
Aim of the experiment: purification of pSB1C3 with RFP-Reportercasette for further processing (P75)
Procedure:
- Minipreps according to the manufacturer's protocol (with 8* 6 mL culture)
- Resulting plasmid: P75
- Concentration: 142.2 ng/µl [~200 µL]
Inoculation of colony from NanoLuc [P54, Repeat]
Investigator: JH
Procedure:
- 5 ml LB+Cam media
- culture was inoculated with one colony
- Incubation at 37°C overnight
Saturday, June 18th
Miniprep of P54 (NanoLuc)
Investigator: LK
Aim of the experiment: Purification of P54 (NanoLuc)
Procedure:
- Miniprep according to the manufacturer's protocol
- Resulting plasmid: pSB1C3 mit NanoLuc (P54, von )
- Concentration: 497,9 ng/µl
Hybridisation of O19 and O20 (StrepTag)
Investigator: LK
Aim of the experiment: Hybridisation of the Strep-Oligos
Procedure: 2µl of each Oligo where diluted in 16µl ddH2O and incubated for 10 min. at 95°. Cooling down at RT for at least half an hour. Stored in fridge
Week 6 (June 20th - June 26th)
Monday, June 20th
Miniprep of F42 (IRES-BirA in pSB1C3)
Investigator: LK
Aim of the experiment: Purification of IRES-BirA Plasmids
Procedure:
- Miniprep according to the manufacturer's protocol
- Resulting plasmid: pSB1C3 with IRES-BirA
Next Steps:
- analytical digestion with EcoRI+PstI and gel (expected bands at 1500 and 2000 Bp)
- Sequencing
Results: Concentration: 583,5 ng/µl
PCR and Purification of BirA
Investigator: LK, JB
Aim of the experiment: Amplification of BirA-construct
Procedure:
- PCR according to the SOP (primers: O17 and O18)
- PCR purification via the purification kit according to the manufacturer's protocol
- Analysis on an 1% agarose gel using 5 µl purified PCR product
Results:
- Concentration: 185,8 ng/µl --> F41
- Analytical gel shows no significant band at 960 Bp -->Repeat
Next Steps:
- Repetition of PCR and analytical gel!
Gel and Gelextraction of P19 (QC EspP) and F28 (EspP and mRuby)
Investigator: LK
Aim of the experiment: Purification of both constructs for Ligation
Procedure:
- Preperative gel according to SOP
- The Purification was done according to the manufacturer's protocol. The EGFR gel-fragment was eluted in 50µl and the mRuby-fragment in 30µl (because of thin band)
Results:
- One thick band just before 1000Bp and one lighter band just before 750 Bp. This fits to the lengths of EspP and mRuby.
- Concentrations: EspP(now F43): 49,5ng/µl mRuby(now F44): 9,9ng/µl
Next Steps:
- Ligation into pSB1C3
Sequencing of F42 (pSB1C3(IRES-BirA)), P54 (pSB1C3(NanoLuc)) , P74 (pSB1C3(RFP generator)) and P76 (pSB1C3(strong RBS-pCat))
Investigator: JL
Aim of the experiment: Sequencing of F42, P54, P74 and P76 in forward direction.
Procedure: Sequencing batches were prepared after manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
The different plasmids we prepared received the following barcodes:
- F42 (VF2): FR11326601
- P54 (VF2): FR11326600
- P74 (VF2): FR11326599
- P76 (VF2): FR11326598
PCR and Purification of genesynthesis 6 (iGEM6) Streptavidin/Traptactin
Investigator: NA
Aim of the experiment: Amplification of gen synthesis 6
Procedure: PCR according to the SOP (primers: VF2 and VR)
Next Steps:
- PCR purification via the purification kit according to the manufacturer's protocol
- Analysis on an 1% agarose gel using 5 µl purified PCR product
- PCR purification via the purification kit according to the manufacturer's protocol
- Analysis on an 1% agarose gel using 5 µl purified PCR product
PCR and Purification of BirA (F41)
Investigator: NA
Aim of the experiment: Amplification of BirA (F29)
Procedure: PCR according to the SOP (primers: O17 and O18)
Next Steps:
- PCR purification via the purification kit according to the manufacturer's protocol
- Analysis on an 1% agarose gel using 5 µl purified PCR product
- PCR purification via the purification kit according to the manufacturer's protocol
- Analysis on an 1% agarose gel using 5 µl purified PCR product
--> repeat with gene-synthesis 2 (F29 may not be the gene-synthesis 2)
Ligations of StrepTag, EspP and mRuby3 with F30 (digested pSB1C3)
Investigator: JH
Aim of the experiment: Ligations of StrepTag (O19+O20), EspP (F43) and mRuby3 (F44) into F30 (NgoMIV and SpeI; pSB1C3 without insert)
Procedure:
- Ligation mixtures were prepared as following:
- O19+O20: 1.7 (O19+O20), 8.3 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- F43: 4.6 μL F43, 5.4 µL F30, 2 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- F44: 8.1 μL F44, 1.9 μL F30, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- Ligation was incubated overnight at 16 °C
Digestion of GSY5 (Streptactin; F27)
Investigator: LK
Aim of the experiment: Restricting GSY5 into StrepActin fragment (Streptavidin mutant) and Leptin fragment (Leptin is for Volker)
Procedure:
- Restriction mix:
32µl DNA (F27)
10µl ddH2O
1µl of each enzyme (SpeI, HindIII, SapI)
5µl cut smart buffer
Next Steps:
- Preperative gel, 1,5% Agarose, cut out and purify bands at 80 Bp and 425 Bp. (80 is leptin for volker and 425 is StreptActin for further cloning into pASK)
Results:
Tuesday, June 21st
Plating of BioBricks that didn't survive sequencing
Investigator: JB
Aim of the experiment: Instead of directly inoculating cultures with the transformed cells from the bacterial agar that was sent by the HQ, the bacterial agar was now used for overnight plating on agar plates containing the appropriate antibiotic resistance.
Procedure: Two methods were used for each BioBrick: For the first method, an inoculation loop was used for plating directly. For the second methods, the inoculation loop was used to inoculate a 50 µl LB culture that
Sequencing of various plasmids
Investigator: CG
Aim of the experiment: Sequencing of various plasmids with either VF2 or VR
Procedure: Refer to the “Inventarliste” for detailed informations on sequencing codes and primers. Sequencing batches were prepared according to manufacturer's protocol (15 µL plasmid DNA (50-100 µM) and 2 µL sequencing primer)
PCR of gene synthesises (iGEM_1) (mRuby_EGFR)
Investigator: CG
Aim of the experiment: iGEM_1_mRuby_EGFR (F28) for preparing the EGFR signal (at about 100 bp)
Procedure: Approach:
- 1µl DNA (diluted, 1:10)
- 25 µl 2x MasterMix
- 0,5µl dNTPs
- 2,5 µl forward Primer VF2
- 2,5µl reverse Primer VR
- Fill up with water up to 50 µl
Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/)
- 98°C_1min initiation step
- 98°C_10sec
- 66°C_30sec > step 2-4: 35 repeats
- 72°C_30sec
- 72°C_2min final step
- 4°_hold
Transformation of Ligations of F50 (StrepTag), F51 (EspP) and F52 (mRuby3) in pSB1C3
Investigator: JH
Aim of the experiment: Transformation of StrepTag (F50), EspP (F51) and mRuby3 (F52) in XL1 blue
Procedure: transformation in competent E. Coli XL1 blue according to protocol + rescue (repeat due to incubation with LB+Cam ! )
Result: plates (LB Cam) in incubator for further processing (37 °C)
Digestion of P9 (pASK75), P74 (pSB1C3), P26 (CMV promotor) and F47 (BirA)
Investigator: JB
Aim of the experiment: Digestion of P9 and P74 to gain the vectors pASK75 and pSB1C3, respectively, as well as digestion of F47 (PCR fragment of BirA) and P26 (CMV promoter) for the ultimate creation of biobricks
Procedure: P74 was digested in a total volume of 30 µl, while the others were digested in a total volume of 50 µl. Enzymes used were:
- P74: XbaI and PstI
- P9: HindIII and XbaI
- F47: Xba I and PstI (didn’t work)
- P26: SpeI and PstI
The digestions were incubated at 37°C for 40 minutes and were thereafter inactivated at 80°C for 5 mins.
Inoculation of cultures for minipreps for P76 (pCat + strong RBS)
Investigator: JB
Procedure: Inoculation of 2x5 ml cultures with P76 clones. Incubation at 37°C overnight.
Wednesday, June 22nd
Minipreps of P79 (pCat+RBS)
Investigator: JB
Procedure: According to the manufacturer's protocol.
Results: Concentration 212 ng/µl. Sent to sequencing
Transformation of ligation F56 (pASK75(Streptactin)), P60 (pSB1C3 (pC+AraC+pBad+GFPc)) , P70 (pSB1C3 (short linker))
Investigator: CG
Aim of the experiment:' Transformation of pASK75-Streptactin (F56) into E. coli XL1 blue, K577881 (P60) into BL21 and K243004 (P70) in XL1
Procedure: transformation in competent E. Coli XL1 blue according to protocol
Result: plates (LB + appropriate antibiotic) in incubator for further processing (37 °C)
Inoculation of colonies from K157001 (EGFR signal peptide), EspP (F50), mRuby (F51), Strep-Tag (F52)
Investigator: CG
Procedure:
- 2x 5 ml LB+Cam (Amp for K157001) media
- Each culture was inoculated with one colony, for ligations 2 colonies each
- Incubation at 37°C overnight
Plateing of K243004 (short linker), K243005 (middle linker) from stab cultures
Investigator: CG
Procedure: For both biobricks: stabing into the agar with the inocluation loop and resuspending in 50 µl LB media. Plating on Amp-agar.
Digestion of P64 (EGFR sgnal peptide) and P71 (CD4 signal peptide) for cloning into F57 (pSB1C3 (CMV promotor))
Investigator: CG
Aim of the experiment: Digestion of P67 and P71 for cloning the resulting fragments into the digested F57 fragement.
Procedure: both plasmids were digested in 50 µl, 3 µg for each plasmid DNA. 1 µl of XbaI and PstI, 5 µl Buffer, 20 µl DNA, 23 µl H2O. The digestions were incubated at 37°C overnight.
Preparative agarose gels for separation of digestion of P64 (EGFR sgnal peptide) and P71 (CD4 signal peptide)
Investigators: NA, JB
Aim of the experiment: Separation of digestion fragments of both digested PCR fragments as well as vectors
Procedure:
- P74: XbaI and PstI -> Fragment corresponding to the pSB1C3 vector isolated
- P9: HindIII and XbaI -> Fragment corresponding to the pASK75 vector isolated
- F47: Xba I and PstI (didn’t work) –> only one fragment at 3 kbp present (weird since that was a gene synthesis pcr)
- P26: SpeI and PstI -> Fragment corresponding to the cmv-promoter isolated
- Gelextraction of P9, P74 and P26 was executed by manufacturers protocol (Qiagen)
Results:
- F55: pASK75 (2,9 ng/µl)
- F56: pSB1C3 (23,6 ng/µl)
- F57: CMV-Promotor (5,9 ng/µl)
PCR of GSY2 (IRES-BirA) with O17/18
Investigators: LK
Aim of the experiment: Amplification of BirA, using new dilution of GSY2 this time, because last 2 PCRs showed confusing results (bands at 2500)
Procedure:
- PCR was performed according SOP
- Primers: O17 and O18
- Annealing at 68°C
Next Steps:
- PCR-Purification
- Analytical Gel
- Ligation into pSB1C3
QuickChange-PCR of pASK75 with O21 and O22
Investigators: LK
Aim of the experiment: Deleting NdeI from pASK (P03)
Procedure:
- QC-PCR was performed according to SOP
- Primers: O-021 and O-022
- Annealing at 68°C
- Elongation time: 3:30 min
Next Steps:
- Digestion with DpnI at 37°C for 1h
- Purification?
- Preperative Gel (band at 960), purification
- Ligation into pSB1C3
Verification of PCR and Digestion of F28 (gene synthesis 1)
Investigator: NA, LK
Aim of the experiment: Verification of successful PCR via gel electrophoresis and preparation of EGFR
Procedure:
- Gel electrophoresis with 1 kb-ladder
- Digestion: 42µl of PCR Purification, 1,5 µl of each enzyme (NgoMIV and SpeI), 5µl of cut smart buffer.
- Incubation at 37°C over night
Results:
Next steps:
- Preparative gel and gel extraction
- Ligation into pSB1C3
Thursday, June 23th
Miniprep of E. coli Xl1-Blue transformed with ligation product P80/81 (mRuby3 K1/2), P82/83 (EspP K1/2), P84/85 (StrepTag K1/2) and Trafo of K157001
Investigator: Jan, Julian
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with ligation product F50(K1,2), F51(K1,2), F52(K1,2) and Trafo of K157001
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P80 | 432,7 |
| P81 | 294,8 |
| P82 | 450,5 |
| P83 | 479,0 |
| P84 | 108,0 |
| P85 | 356,0 |
| P86 | 47,2 |
PCR of gene synthesises
Investigator: CR
Aim of the experiment: iGEM_1_mRuby_EGFR (F28) for preparing the EGFR signal (at about 100 bp)
Procedure: Approach:
- 1µl DNA (diluted, 1:10)
- 25 µl 2x MasterMix
- 0,5µl dNTPs
- 2,5 µl forward Primer VF2
- 2,5µl reverse Primer VR
- Fill up with water up to 50 µl
Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/)
- 98°C_1min initiation step
- 98°C_10sec
- 66°C_30sec > step 2-4: 35 repeats
- 72°C_30sec
- 72°C_2min final step
- 4°_hold
QuickChange-PCR of pASK with O21 and O22
Investigators: CR
Aim of the experiment: Digestion of PCR product with DpnI to digest methylated DNA
Procedure:
PCR purification of BirA and mRuby/EGFR
Investigator: CR
Aim of the experiment: purification of PCR
Procedure: PCR purification via the purification kit according to the manufacturer's protocol The samples were renamed:
- BirA: F59
- mRuby/EGFP: F60
The samples were analysed on a 1% agarose gel. Samples were prepared after SOP.
Results:
Transformation of P60 (K577881; pC+AraC+pBad+GFPc K1) and QC P3 (pASK75(SA1))
Investigator: EF, JB
Aim of the experiment: Transformation of Q CP3 in XL1 blue; transformation of p60 in BL21
Procedure: transformation in competent E. Coli BL21 and XL1blue according to protocol
1µl p60 in BL21 7 µl Q CP3 in XL1 blue
Result: plates (+ rescue plates) incubating
Cloning of CD4- and EGFR-signal peptide into pSB1C3-CMV
Investigator: CR
Aim of the experiment: Digestion of P71 and P69 Procedure:
- Digestions were prepared as following:
- 20 µl CD4 (P71); 1 µl XbaI; 1 µl PstI; 5 µl CutSmart; 23 µl ddH2O
- 7 µl pSB1C3-CMV; P26; 1 µl PstI; 1 µl SpeI; 2 µl CutSmart; µl ddH2O
- 20 µl EGFR-signal peptide (P67);1 µl XbaI; 1 µl PstI; 5 µl CutSmart; 23 µl ddH2O
- Digestions were incubated for 3 h at 37 °C
- The Samples CD4- and EGFR-signal peptide were applied to a 2% and the pSB1C3-CMV to a 1% agarose Gel
Results:
--> passt
--> passt
Next steps:
- Ligation of
Cloning of EGFR (F62) / CD4 (F61) signale peptide into pSB1C3-CMV (F63)
Investigator: CG
Aim of the experiment: Gelextraction and ligation of EGFR- and CD4-signal peptide into pSB1C3 and transformation
Procedure:
- Gel extraction according to manufacturers protocol (Qiagen): F61 (CD4) 2 ng/µl, F62 (EGFR) 0.6 ng/ µl, F63 (CMV): 45 ng/ µl
- Ligation: 1 µl T4 Ligase, 2 µl Ligase buffer, 7 µl H20, 1.8 µl vector and 8.2 µl EGFR insert or 4.3 µl vector and 5.7 µl CD4 insert
- Transformation according to SOP
- Plating on LB-Cam agar
Friday, June 24th
Analytical digestion and gelelectrophoresis of P83 (EspP K2) and P83 (StrepTag K2)
Investigator: Julian, Niklas, Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P82/83 (EspP K1/2) and P84/85 (StrepTag K1/2).
Procedure:
- Batch for analytical digestion for P82-P85 with EcoRI-HF
| volume | reagent |
| 0.5/1.0 µl | Plasmid DNA (-/P84) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) |
| 8/7.5 µl | ddH2O (-/P84) |
| =10 µl | TOTAL |
Sequencing of P80( mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Investigator: Julian
Aim of the experiment: Sequencing of P80(mRuby3 K1), P83 (EspP K2) and P85 (StrepTag K2)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). Sequencing primer VF2 was used
The different vectors we sequenced received the following barcodes:
- mRuby3 in pSB1C3 (P80): FR11326590
- EspP in pSB1C3 (P83): FR11326588
- Streptag in pSB1C3 (P85): FR11326587
Transformation of E. coli XL1 blue with F64 (quickchanged P3(pSAm1))
Investigator: Niklas
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C.
Gelextraction of F67(BirA), F68(mRuby), F69(EGFR-TMD), F70(pSB1C3) and F71(pSB1C3)
Investigator: Niklas
Aim of the experiment: Gelextraction of F67(BirA(Digest. F59 [EcoRI; SpeI])), F68(mRuby(Digest. F60 [NgoMIV; SpeI])), F69(EGFR-TMD(Digest. F60 [NgoMIV; SpeI]), F70(pSB1C3(digest. P74 [NgoMIV; SpeI]) and F71(pSB1C3(digest. P74 [EcoRI; SpeI])
Procedure:
Gelextraction was performed by manufacturers protocol (Qiagen).
Cloning of F59 (BirA) and F60 (EGFR-TMD) into pSB1C3
Investigator: CR
Aim of the experiment: Generate biobricks with EGFR-TMD and BirA
Procedure:
- Digestions were prepared as following:
- 1. xµl F59 (BirA); 1 µl EcoRI; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O
- 2. 2 µl P74 (pSB1C3 (RFP-generator); 1 µl NgoMIV; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O
- 3. xµl F60 (mRuby/EGFR); 1 µl NgoMIV; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O
- 4. 2 µl P74 (pSB1C3 (RFP-generator), 1 µl EcoRI; 1 µl SpeI; 3 µl CutSmart; 23 µl ddH2O
- Digestion was incubated for 3 h at 37 °C
To Do:
- Gel (already prepared)
- Gelextraction
- Ligation
- Trafo
Results:
1. band: P74(NgoMiV, SpeI) 2. band: P74 (EcoRI, SpeI)
1. band: F59 2. band: F60
Transformation of EGFR-TMD-pSB1C3 and BirA-pSB1C3 ligations
Investigator: JB
Procedure: According to protocol. Plates were incubated overnight at 37°C.
Chemical biotinylation of BSA
Investigator: MT, JB
Procedure: BSA was chemically biotinylated with a 20x, 40x and 60x molar excess in two different buffers:100 mM Bicin (pH 8.85) and 100 mM borate (pH 8.85) with 50 mM NaCl respectively.
-> A total of six different tubes.
After one hour incubation at RT, the samples were put in the freezer
Inoculation of liquid cultures (F65 and F66)
Investigator: JB
Procedure:
- Clones were picked for inoculation of 5 ml liquid cultures containing the appropriate antibiotic
- Incubation overnight at 37°C/shaking
Saturday, June 25th
Miniprep of E. coli Xl1-Blue transformed with P60 (mRuby/EGFR),F58 (Ligation pASK75 + Streptactin), F65 (CMV + CD4), F66 (CMV + EGFR), P70 (Short Linker)
Investigator: Niklas
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P87 | 81 |
| P88 | 34,5 |
| P89 | 86,3 |
| P90 | 108,5 |
| P91 | 417,4 |
Analytical digestion and gelelectrophoresis of F64 (quickchanged P3, pASK75(SA1)) for verification of success of quickchange
Investigator: Niklas
Procedure:
- Analytical digestion with NdeI and gelelectrophoresis. If quickchange worked there should be a band at about 3200 bp (only one restriction site left)
- Incubation over night at room temperature.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
Just a band showing a few bp (Primer), there is no plasmid band -> Quickchange did not work
Inoculation of colonies from Ligation of F69 + F70 (EGFR-TMD in pSB1C3) and F44 + F33 (mRuby in pSB1C3)
Investigator: Niklas
Procedure:
- 6x 4 ml LB+Cam media
- Each culture was inoculated with one colony
- Incubation at 37°C overnight
Sunday, June 26th
Miniprep of E. coli Xl1-Blue transformed with ligation product F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3)
Investigator: Luisa
Aim of the experiment: Extracting F44 + F30 (mRuby3 in pSB1C3), F66 + F70 (CMV+EGFR in pSB1C3) from E.coli XL-1-blue
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P92 | 162,9 |
| P93 | 447,9 |
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- digestion of PCR-Product with DpnI for 1h at 37°C
- now labeled P94
Transformation of E. coli XL1 blue with P94 (quickchanged P3(pSAm1))
Investigator: Luisa
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) for pASK (F72) and on chloramphenicol plates for P92 and P93 and incubated over night at 37 °C in the incubator.
Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75)
Investigator: Luisa
Aim of the experiment: Analytical digestion and gelelectrophoresis of P92 (mRuby), P93 (CMV + EGFR) and P94 (pASK75).
Procedure:
- Batch for analytical digestion for P92 and P93 with EcoRI-HF and PstI-HF
| volume | reagent |
| 1 µl | Plasmid DNA |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) and PstI-HF (10 U/µl) for P92/93, NdeI (10 U/µl) for P94 |
| 7.5/7 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- 1. band (P92): no mRuby at 700bp visible, only empty vector
- 2. band (P93): empty vector and EGFR --> perfect
- 3. band (P94): no signal at all --> repetition of QC-PCR
Week 7 (June 27th - July 3rd)
Monday, June 27th
Sequencing of P67 (EGFR-Signalpeptid)
Investigator: Niklas
Procedure:
Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2))
FR11326586
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- Digestion of PCR-Product with DpnI for 1h at 37°C.
- Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR).
PCR of Genesynthesis 3 and 4
Investigator: Luisa
Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)
Procedure:
- The PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 2,5 µl | Primer VF2 |
| 2,5 µl | Primer VR2 |
| 1 µl | dNTP-mix |
| 10 µl | Q5 Polymerase reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Q5-Polymerase |
| 18 µl | ddH2O |
- Setup: iGEM_standard (Promega-cycler)
| temperature | time |
| 98°C | 2min |
| 98°C | 10sec |
| 66°C | 30sec |
| 72°C | 30sec |
| 72°C | 2min |
| 4°C | hold |
- the batches were then purified using the Quiagen PCR-Purification Kit.
Analytical digestion and gelelectrophoresis of P88 (pASK75(Steptactin)), P89 (pSB1C3(CMV+CD4) and P90 (pSB1C3(CMV+EGFR-Signal peptide)
Investigator: Niklas
Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)
Procedure:
- Batches for analytical digestions:
P88: EcoRI
P89: EcoRI and PstI
P90: EcoRI
| volume | reagent |
| 5,8/2,3/1,8 µl | Plasmid DNA (P88/P89/P90) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl)/ PstI |
| required amount for total volume of 10 µl | ddH2O |
Ligation of F67 (BirA) and F71 (vector fragment pSB1C3), Transformation of E. coli XL1 blue afterwards
Investigator: Niklas
Aim of the experiment: Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 2,4 µl | Vektor |
| 7,6 µl | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
- next step: analytic digestion of transformation was successful
Digestion of PCR on genesynthesis 3 and 4, and pSB1C3
Investigator: Luisa
Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.
Procedure:
- Batches for analytical digestions:
| volume | reagent |
| 1 µl each | enzyme (SapI, HindIII, XbaI, AgeI) |
| 5 µl | CutSmart buffer (10x) |
| 41 µl | DNA (purified PCR-products of GSY3 and 4) |
- Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80.
Preparative digestion and gelelectrophoresis of P80 , P78 and P85
Investigator: Julian
Aim of experiment: Preparative digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)
Procedure:
- Batches for analytical digestions:
- P80 and P78: EcoRI and AgeI
- P85: EcoRI and NgoMIV
| volume | reagent |
| 10 µl | Plasmid DNA |
| 32 µl | ddH2O |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme(10 U/µl)/ PstI |
| 50 µl | TOTAL |
Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into E. coli XL1 blue
Investigator: Luisa
Aim of the experiment: Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 4,3µl(for F75), 8,2µl (for F76) | Vector |
| 12,7µl (F75), 8,8 (F76) | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| =20 µl | TOTAL |
- Ligation was incubated at RT for 1,5h.
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator.
Chemical biotinylation of BSA
Investigator: Niklas
Procedure:
- BSA was chemically biotinylated with a 20x and 40x molar excess:
- 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85)
- dissolve BSA (10 mg/ml)
- Add biotin-NHS-ester: 20,5 mg for 40x molar excess
- reaction over night
Tuesday, June 28th
Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator)
Investigator: Julian
Aim of experiment: Preparative digestion and gelelectrophoresis of P83 (EspP) and P29 (DoubleTerminator)
Procedure:
- Batches for analytical digestions:
- P83: EcoRI and SpeI
- P29: EcoRI and XbaI
| volume | reagent |
| 6.5/13 µl | Plasmid DNA (P83/P29) |
| 35.5/29 µl | ddH2O (P83/P29) |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme |
| 50 µl | TOTAL |
- Preparative gelelectrophoresis was performed at 80 V for 80 min
Gelelectrophoresis of F73 (pSB1C3 digst. EcoRI, AgeI)
Investigator: Julian
Aim of experiment: gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85 (Strep-Tag)
Procedure: Preparative gelelectrophoresis was performed at 100 V for 45 min using a pocket spanning over the full width of the 1% agarose gel
Wednesday, June 29th
Transformation of F83 (EspP-Fragment in Double Terminator + pSB1C3), F84 (GSY3 + pSB1C3) F85 (GSY4 + pSB1C3)
Investigator: Niklas
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
Thursday, June 30th
Transformation of new E. coli XL1-blue (P75), additional plating of untransformated XL1-blue
Aim of experiment: Quality assurance
Investigator: Niklas
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
- additional plating of untransformated XL1-blue on Tet-, Cam-, Amp- and Kan-plates
Result:
- After 12 hous of incubation there were some (~20) colonies on the 50µl-plate transformed with Cam-resistence-plasmids. The transformation has been succesful, though cells grow slow.
- There were no colonies on the kan-, amp- and cam- plates with the untransformed E.Coli. Hence we can assume that there are no other plasmids with resistences against kan, amp or cam in our batch of competent cells.
There are cells on the tet-plate. This was expected.
Friday, June 31th
Transformation of P9 (pASK with Streptavidin WT) and QC-PCR product into new E. coli XL1-blue
Aim of experiment: Tranforming pASK without NdeI-cutting point into E.coli (two batches from diffrent approches were tested). And Transforming pASK_SA1 into E.coli XL-1-blue.
Investigator: Luisa
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 30 sec. heat shock at 42 °C
- Adding of 950 µl LB-medium to each tube. 1h hour incubation at 37°C.
- The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C.
Week 8 (July 04–10)
Monday, July 04
Repetition of the Transformation of QC-PCR product into new E. coli XL1-blue
Aim of experiment: Tranforming pASK without NdeI-cutting point into E.coli (two batches from diffrent approches were tested).
Investigator: Luisa
Procedure:
- the new competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 30 sec. heat shock at 42 °C
- Adding of 950 µl LB-medium to each tube. 1h hour incubation at 37°C.
- The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C.
Transformation of P9 (Streptavidin in pASK75) into E. coli BL-21
Aim of experiment: Tranforming pASK with Streptavidin into E.coli BL-21 for Expression.
Investigator: Niklas
Procedure:
- the new competent E. coli BL-21 cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cells were plated out on Amp-plates (inclusive rescue plate) and incubated over the weekend at 30 °C
Transformation of F94 (hGH polyadenylation sequence and Nanoluc with StrepTag)
Investigator: Javier
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 45 sec. heat shock at 42 °C
- Adding of 950 µl LB-medium.
- The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
Miniprep of E. coli Xl1-Blue transformed with ligation product P108 and P113 (EspP-Fragment in Double Terminator, F81+F82), P109 and P110 (GSY3+ F73 vector AgeI+XbaI ), P111 and P112 (GSY4+ F73 vector AgeI+XbaI)
Investigator: Javier
Aim of the experiment: Extracting P108 and P113 (EspP-Fragment in Double Terminator, F81+F82), P109 and P110 (GSY3+ F73 vector AgeI+XbaI ), P111 and P112 (GSY4+ F73 vector AgeI+XbaI) from E.coli XL-1-blue
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P108 | 116,2 |
| P109 | 86,6 |
| P110 | 136,8 |
| P111 | 144,8 |
| P112 | 163,2 |
| P113 | 123,3 |
Preparative digestion and gelelectrophoresis of P74 (RFP-Gen.)
Investigator: Julian
Aim of experiment: Preparative digestion and gelelectrophoresis of P74 (RFP-Gen.)
Procedure:
- Batch for analytical digestion:
- P74: EcoRI and SpeI
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 39.5 µl | ddH2O |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme |
| 50 µl | TOTAL |
- Preparative gelelectrophoresis was performed at 90 V for 45 min
Result: pSB1C3 (EcoRI und SpeI) F95
Cloning of F96 (BirA in pSB1C3)
Investigator: Julian
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 5 min incubation on ice
- 30 s heat shock at 37 °C
- Adding of 950 µl LB-medium
- Incubation at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C
Tuesday, July 05th
Preparing of 200 ml TB medium for inoculation of 2 L medium on Thursday
Investigator: Niklas
Procedure:
- 2.4 g Tryptone 4.8 g yeast extract
- 800 µl Glycerine
- adjust to 180 ml with ELGA-water
- seperately: 3.29 g H2KPO4 + 0,46 g K2HPO4 with ELGA water (Volume = 20 ml)
Digestion of F49 (BirA) with EcoRI and SpeI and cloning in F95 (pSB1C3)
Investigator: Clemens
Procedure:
- Digestion of F49 with EcoRI and SpeI
| volume | reagent |
| 20 µl | Plasmid DNA (F49) |
| 5 µl | CutSmart buffer (10x) |
| 1 µl | EcoRI-HF(10 U/µl) |
| 1 µl | SpeI(10 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Digestion was incubated for 1.5 h at 37°C
- Digestion was applied to a 1% agarose-gel
- Gel ran at 80V for 35 min
File:Muc16 digestF49 0507.jpeg
- BirA had a length of 1012 bp
- Upper probe was cut from the gel
- BirA was purified with gelextraction which was performed by manufacturer's protocol
- Concentration was measured
- [F97]= 12.4
- Ligation of F97 into pSB1C3 (F95)
| volume | reagent |
| 1.2 µl | Vector: pSB1C3 (F95) |
| 8.8 µl | Insert: BirA (F97) |
| 2 µl | T4 DNA Ligase buffer |
| fill up to 20 µl | dd H2O |
| 1 µl | T4 ligase |
| =21 µl | TOTAL |
- Ligation was incubated at RT for 30 min
- Transformation:
- Transformation was performed after SOP
- 7 µl were used for transformation in E. coli XL1blue
Sequencing of P108 - P112
Aim of experiment: Sequencing
Investigator: Niklas
Procedure:
- sequencing batches after protocol with following barcodes:
- P108 (EspP-Fragment in Double Terminator (F83)) FR11326572 (VF2)
- P109 (GSY3+ F73 Leervektor AgeI+XbaI (F84)) FR11326576 (VF2)
- P110 (GSY3+ F73 Leervektor AgeI+XbaI (F84)) FR11326575 (VF2)
- P111 (GSY4+ F73 Leervektor AgeI+XbaI (F84)) FR11326574 (VF2)
- P112 (GSY4+ F73 Leervektor AgeI+XbaI (F84)) FR11326573 (VF2)
NEB Turbo CaCl2-competent cells
Investigator: Vivien
Procedure:
- Inoculate 50 mL LB with 0.5 mL overnight culture of NEB Turbo cells
- Incubate at 37 °C for 7 h; final OD500: 0.357
- Very little growth in the course of the last hour; OD500 0.45 required
- Experiment discontinued. Inoculated 3 further colonies in 5 mL LB for o/n growth (37 °C). Repeat experiment on Wed, July 6
Miniprep P114 (NanoLuc + StrepTag + Poly-A in pSB1C3(F94))
Investigator: Niklas
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of P114 and P115
Investigator: Niklas
Aim of the experiment:
- Analytical digestion and gelelectrophoresis of P114 and P115 (NanoLuc + StrepTag + Poly-A in pSB1C3 K1 and K2)
Procedure:
- Batch for analytical digestion for P114 and P115 with EcoRI+PstI
| volume | reagent |
| 1.75 µl | Plasmid DNA P114/P115 |
| 1 µl | cut smart (10x) |
| 0.5 µl | EcoRI |
| 0.5 µl | PstI |
| 6,25 µl | ddH2O |
| =10 µl | TOTAL |
- Incubation for 45 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
File:Muc16 digestP114 P115 0507.jpeg
| 1 kbp ladder DNA ladder | P114 | P115 |
| X | X |
Inoculation of Quick change of P88 (pASK75 (Streptactin)) preculture
Investigator: Clemens
Procedure:
- four colonies were picked from plate
- Cells were inoculated in 5 mL LB medium (ampicillin 1:1000)
Analytical digestion of P108 and P113 (both EspP + double terminator (F83)) with EoRI and PstI
Investigator: Clemens
Procedure:
- Digestion of F49 with EcoRI and SpeI
| volume | reagent |
| 1 µl | Plasmid DNA (P108/P113) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl) |
| 0.5 µl | PstI(10 U/µl) |
| 7 µl | ddH2O |
| =10 µl | TOTAL |
- Digestion was incubated for 1.5 h at 37°C
- Digestion was applied to a 1% agarose-gel
- Gel ran at 80V for 35 min
File:Muc16 analyt.digest.P108 P113 0507.jpeg
- P108 and P113 showed the right bands
Wednesday, July 06th
Sequencing of P108 (EspP-Fragment in Double Terminator (F83))
Investigator: >Niklas
Procedure:
- sequencing batches after protocol with following barcodes:
- P108 (EspP-Fragment in Double Terminator (F83)) FR11326572 (VR)
Digestion of P75 (RFP-Generator) with HincII + dephosphorylation
Investigator: Niklas
Aim of experiment: Blunt-Ligation of BirA into pSB1C3
Procedure:
- Digestion of P75 (RFP-Generator) with HincII + dephosphorylation
| volume | reagent |
| 3 µl | Plasmid DNA (P75) |
| 2 µl | CutSmart buffer (10x) |
| 0.5 µl | HincII |
| 1 µl | Fast AP (for dephosphorylation) |
| 14.5 µl | ddH2O |
| =20 µl | TOTAL |
- Digestion was incubated for over night at 37°C
- next steps: PCR of GSY2 with O17 and O18 + phosphorylation, Ligation in digested pSB1C3 afterwards
Cloning of A3C5 Tag into the MiddleLinker-Plasmid
Investigator: Christoph
Aim of experiment: Cloning of A3C5 Tag into the MiddleLinker-Plasmid
Procedure:
- Digestion of Middle_Linker Plasmid (P69) with AgeI and PstI
| volume | reagent |
| 0.3 µl | Plasmid DNA (69) |
| 2 µl | CutSmart buffer (10x) |
| 1 µl | AgeI |
| 1 µl | PstI |
| 1 µl | Fast AP (for dephosphorylation) |
| 14.5 µl | ddH2O |
| =20 µl | TOTAL |
| volume | reagent |
| 0.3 µl | Plasmid DNA (P112) |
| 2 µl | CutSmart buffer (10x) |
| 1 µl | NgomIV |
| 1 µl | PstI |
| 14.5 µl | ddH2O |
| =20 µl | TOTAL |
- Digestion was incubated for over night at 37°C, heat inactivation at 80°C for 20 min.
| volume | reagent |
| 2 µl | digested Middle-Linker (F99) |
| 8 µl | digested A3C5 (F100) |
| 2 µl | 10x Ligase Buffer |
| 1 µl | Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- Transformation according to SOP in E. coli XL1 blue
Picking of of E. coli XL1 blue with Streptactin in quickchanged pASK75, F84 (digested GSY3), F85 (digested GSY4)
Investigator: Niklas
Procedure:
- Streptactin in quickchanged pASK75: Colonies were picked from Amp-plates. (10x)
- F84/F85: Colonies were picked from Cam-plates. (8x each)
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL of the proper antibiotic.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Quick Change of mRuby Plasmids
Investigator: Christoph
Aim of experiment: Correction of wrong amino acid within the mRuby sequence
Procedure:
QC according to manufacturers protocol
| volume | reagent |
| 10 ng (= 0,3 µl 1:10) | Plasmid DNA (80) |
| 5 µl | 10x Pfu Ultra II Buffer |
| 1.25 µl | O23 |
| 1.25 µl | O24 |
| 0.5 µl | Pfu Ultra II |
| 1 µl | dNTPs |
| 40.7 µl | ddH2O |
| =50 µl | TOTAL |
| volume | reagent |
| 10 ng (= 0,3 µl 1:10) | Plasmid DNA (100) |
| 5 µl | 10x Pfu Ultra II Buffer |
| 1.25 µl | O23 |
| 1.25 µl | O24 |
| 0.5 µl | Pfu Ultra II |
| 1 µl | dNTPs |
| 40.7 µl | ddH2O |
| =50 µl | TOTAL |
- PCR: 2 min. 98°C initial denaturation, 10 sec. 98°C, 30 sec. Annealing st 60 °C, 5 min 68°C, 35 Cycles
- 1 h at 37°C DpnI Digestion
- Transformation according to SOP in E. coli XL1 blue, QC on mRuby-Strep (F103) was also transformed in E. coli Turbo cells for further investigating the growth rate
Miniprep of Quick Change of pASK75-Streptactin + analytical digestion with Nde1
Investigator: Jan
Procedure:
- Miniprep using the kit according to the manufacturer's protocol.
- Although cultures seemed to have grown properly, very low and unreliable DNA concentrations were obtained via Nanodrop measurement and no fragment at all could be detected via analytical agarose gel electrophoresis after 90 minutes digestion with Nde1
Creation of competent NEB Turbo cells
Investigator: Jan
Procedure:
- 250 µl of precultures were used to inoculate 50 ml LB-medium
- After an OD=0.45 was reached, cells were put on ice and made competent according to the manufacturer's protocol
- 50 µl aliquots of competent cells were frozen in liquid nitrogen and stored at -80°C until further use
Inoculation of a BL21 pBAD-GFP (P60) preculture
Investigator: Jan
Procedure:
- A single clone was picked in order to inoculate 5 ml LB medium (+Cam)
- Culture was grown overnight at 37°C
Inoculation of a BL21 preculture for Streptavidin production (P8)
Investigator: Jan
Procedure:
- A clone was picked for the inoculation of 5 ml LB medium
- After 6-8 hours of incubation, the 5 ml preculture was used for the inoculation of a 50 ml LB preculture
Thursday, July 07
PCR Amplification of birA (GSY 2)
Investigator: Vivien
Aim: Amplify birA gene
Procedure:
- Prepared PCR mix (25 μL reaction volume): NEB Q5 High-Fidelity 2× Master Mix (12.5 μL), ddH2O (9.5 μL), F29 (1 μL), O17 and O18 (1 μL each)
- Cycling protocol: start 1.5 min, 98 °C; 35 cycles 30 s, 98 °C – 45 s, 72 °C – 30 s, 72 °C; final elongation 5 min, 72 °C; end 4 °C
- Purified via Quiagen PCR Purification kit following manufacturer's instructions. DNA eluted from column by adding 50 μL EB buffer and heating at 37 °C for 5 min prior to centrifugation.
Results: DNA concentration: 82.7 ng/μL. Stored as F102 at –20 °C
Induction of Streptavidin
Investigator: Vivien
Aim: Induce streptavidin production
Procedure:
- Inoculated 2 L TB medium with 50 mL o/n pre-culture and added 2 mL ampicillin (10 mg/mL in 50% EtOH)
- Let grow (37 °C, 200 rpm) until OD500=0.622 (c. 2.5 h)
- Induced with anhydrotetracycline (1:10,000 i.e. 200 μL of 2 mg/mL stock)
- Let grow (37 °C, 200 rpm) for 4 h
Results: Induced culture for harvesting (see below)
Production of eGFP
Investigator: Clemens, Vivien, Christoph
Aim: Produce eGFP
Procedure:
- Inoculated 50 mL LB+Cam medium with 0.5 mL o/n pre-culture
- Let grow (37 °C, 200 rpm) until OD500=0.58
- Induced with 0.1 mM arabinose (i.e. 100 μL of 50 mM stock)
- After 7 h of induction the cells were stored in the fridge overnight for maturation of the GFP-chromophore
Results:
Harvesting of Streptavidin
Investigator: Niklas
Aim of the experiment: Recombinant expression and purification of streptavidin
Procedure:
- After expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor)
- The supernatant was discarded, and the pellet was transferred into a Falcon and frozen
- next steps: resuspending and homogenizing in the PANDA
Friday, July 08
Analytical Digest of mRuby3 Constructs
Aim of experiment: Verify integration of mRuby3 constructs in F103 and F104
Investigator: Vivien
Procedure:
- Mixed 16 μL F103 (or 8 μL F104 plus 8 μL ddH2O) with 2 μL 10× CutSmart buffer and 1 μL each of EcoRI-HF and PstI-HF (20 μL total volume)
- Incubated for 45 min at 37 °C
- Subjected to gel electrophoresis (1% agarose, 80 V, 120 min)
Results: No band around 700 bp as expected for mRuby3 constructs
Picking clones and inoculating cultures for F101 and F103
Investigator: Jan
Procedure:
- All clones available (only two for F101, one for F103) were picked and used to inoculate 5 ml of LB medium containing the appropriate antibiotic
- Cultures were incubated at 37°C overnight
Solubilization, lysis and unfolding of streptavidin
Investigator: Jan
Procedure:
- According to protocol -> lysis of pellet (19.2 g) in 58 ml lysis buffer (50 mM Tris/HCl pH 8, 1 mM EDTA)
- As only two liters of medium were used, the Panda couldn't be utilized for cell lysis -> French Press was used
- Cells were additionally submitted to ultrasonic treatment
- Centrifugation of cells, casting away the supernatant and resolubilizing the pellet in 5 ml GdmCl (6M, pH 1.5)
Sequencing of P88
Aim of experiment: Verification of QC
Investigator: Julian
Procedure:
Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (PR1))
FR11326565
Transformation of P75 (RFP-Generator), F101 (middle linker+ A3C5), P125 (dig. GSY4) into E. coli XL1-blue and P75 in NEB Turbo E. coli
Investigator: Niklas, Vivien
Procedure:
- the competent E. coli cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1 µl of DNA (6 µl for Ligation F101) was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- 30 min incubation on ice
- Adding of 950 µl LB-medium to each tube.
- The cells were plated out on Cam-plates/Amp-plate for F101 (inclusive rescue plate) and incubated over the weekend at 30 °C
QuickChange-PCR of P80 (O23 and O24) and P100 (O19 and O20)
Investigators: Niklas
Aim of the experiment: Changing of amino acid
Procedure:
- the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added)
- Annealing at 55°C
| volume | reagent |
| 1.25µl | O23/O24 and O19/O20 |
| 0,5 µl | dNTP-mix |
| 1 µl | 1:100 P80 / P100 |
| 2.5 µl | Pfu Ultra II-buffer |
| 0.5 µl | Pfu Ultra II Polymerase |
| 19.25 µl | dd H2O |
| 4 x 25 µl | TOTAL |
Preperative gelelektrophoresis of F102 (PCR of BirA) + phosphorylation and ligation into pSB1C3
Investigator: Luisa
Aim of experiment: Blunt-Ligation of BirA into pSB1C3
Procedure:
- The whole PCR-product was applied to a 1% agorose gel. After runnig the gel two lines where visible: one at 1000Bp and one at ~500Bp.
- The 1000 Bp band was cut out and purified with the Gelextraction kit according to manufacturer's protocoll. Concentration: 41,5ng/µl.
- The DNA was phosphorylated as follows:
| volume | reagent |
| 15 µl | DNA (~1pmol) |
| 2 µl | Ligase buffer (10x) |
| 1 µl | T4 Polynukleotide-Kinase (for phosphorylation) |
| 2 µl | ddH2O |
| =20 µl | TOTAL |
- The batch was incubated for 20 min at 37°C and then the enzyme was inactivated for 10 min at 75°C.
- The phosphorylated DNA was then ligated into prepared (HincII-digested) pSB1C3 as follows:
| volume | reagent |
| 2,3 µl | Vector |
| 7,7 µl | Insert (BirA) |
| 2 µl | Ligase Buffer (10x) |
| 1 µl | T4 Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- next steps: Transformation into E.coli Xl-1-blue.
Digestion of P78, P123 (for BAP-Nanoluc-construct),P122 (IgKappa for CMV-IgKappa-construct), P89, P114 (for CMV-EGFR-Nanoluc-StrepTag-PolyA-construct)
Investigator: Luisa
Aim of experiment: Getting of BAP-Nanoluc-construct, CMV-IgKappa-construct and CMV-EGFR-Nanoluc-StrepTag-PolyA-construct.
Procedure:
- DNA was digested according to the following table:
| volume | reagent |
| 3 µg (for P78, P123 and P122) and 1,5 µg (for P89, P114) | DNA |
| 5 µl | CutSmart buffer (10x) |
| 1,5 µl of NgoMIV and PstI | for P78 |
| 1,5 µl of AgeI and PstI | for P123 |
| 1,5 µl of XbaI and PstI | for P122 |
| 1,5 µl of AgeI and PstI | for P189 |
| 1,5 µl of NgoMIV and PstI | for P123 |
| up to 50µl | ddH2O |
| =50 µl | TOTAL |
Results: Concentrations: P78 now F106: 2,7 ng/µl, P89 now F107: 14,4 ng/µl. Gelextractions of P114, P122 and P123 didn't contain DNA. (To few DNA available)
- next steps: Retransformation of verified clones (P114, P89) into Xl-1-blue for amplification of plasmid.
Repetition of digestion of P122 and P123 with more DNA (10µg!), then Gel with 100Bp ladder!
Repetition of digestion of P123 (BAP for BAP-Nanoluc-construct) and P122 (IgKappa for CMV-IgKappa-construct)
Investigator: Luisa
Aim of experiment: Getting of BAP-Nanoluc-construct, CMV-IgKappa-construct
Procedure:
- DNA was digested according to the following table:
| volume | reagent |
| 10 µg → 20µl | DNA |
| 5 µl | CutSmart buffer (10x) |
| 1,5 µl of AgeI and PstI | for P123 |
| 1,5 µl of XbaI and PstI | for P122 |
| up to 50µl | ddH2O |
| =50 µl | TOTAL |
- Incubation at 37°C over night.
- next steps: Gel with 100Bp ladder and Gelex. Then Ligation with prepared vectors from former digestion (F106 an F63)
Week 9, July 11–17
Monday, July 11
MiniPreps of pSb1C3-Middle Linker+A3C5, -BM40, -CMV-EGFR Retrafo, Streptactin in QC pASK75
Investigator: Christoph
Aim of the experiment: Plasmid preparation, analytic digestion, sequencing
Procedure:
- MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Analytic digestion with 0.25 µL EcoRI, 0.25 µL PstI (HF) for pSB1C3 plasmids and 0.5 µl NdeI for pASK75-Streptactin, 1 µL SmartCut Buffer, 3 µL plasmid DNA, 5.5 µL H2O
- 5 µL on 1% agarose gel electrophoresis
- plasmid assignment was tricky because of confusion during the weekend Minipreps
Results:
- pASK75-Streptactin clone 1 and 2 were not visible
- verified Plasmid names are available after sequencing
- Lane 1: 5 µl 1 kb Ladder
- Lane 2: 5 µL Plasmid F101 K1 (seems to be pSB1C3-RFP-Generator)
- Lane 3: 5 µL Plasmid F101 K2 (seems not to be pSB1C3-RFP-Generator)
- Lane 4: 5 µl 100 bp Ladder
- Lane 5: 5 µL P125 K1 (seems not to be pSB1C3-RFP-Generator)
- Lane 6: 5 µL P125 K2 (seems to be pSB1C3-RFP-Generator)
Preperative digestion of NanoLuc + StrepTag + Poly-A (F112) and cloning behind CMV-EGFR (F107)
Investigator: Christoph, Niklas
Aim of the experiment: preperative digestion for cloning of the construct behind of CMV-EGFR
Procedure:
- 15 µl P115 (NanoLuc + StrepTag + Poly-A), 1 µl NgoMiV, 1 µl PstI, 3 µl CutSmart buffer, 10 µl H2O
- Incubation for 2 h at 37°C
- 1% gel electrophoresis
Photo
- gel extraction according to SOP
- Ligation of 2 µl F107 (AgeI, PstI) + 8 µl F112 (NgomIV, PstI) with 2 µl Ligase buffer, 7 µl ddH2O, 1 µl T4-Ligase for 40 minutes
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate).
Cloning of Nanoluc behind BAP
Investigator: Christoph, Niklas
Aim of the experiment: Cloning of Nanoluc behind BAP based (testing fast cloning strategy)
Procedure:
- 5 µl P78 (Nanoluc), 0.5 µl NgoMiV, 0.5 µl PstI, 1 µl CutSmart buffer, 2.5 µl H2O, 0.5 SacI = F108
- 4 µl P123 (pSB1C3-BAP), 0.5 µl AgeI, 0.5 µl PstI, 1 µl CutSmart buffer, 3 µl H2O, 1 µl FastAP = F109
- Incubation for 2 h at 37°C, 15 min heat inactivation
- Ligation: 2 µl F108 (Nanoluc) + 8 µl F109 (BAP), 2 µl buffer, 1 µl Ligase, 7 µl H2O
- Transformation:
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate).
Gel extraction of IgKappa (F111) and cloning behind CMV in pSB1C3 (F63)
Investigator: Christoph, Niklas
Aim of the experiment: gel extraction after digestion for cloning of the construct behind of CMV
Procedure:
- 1% gel electrophoresis
Photo
- gel extraction according to SOP
- Ligation of 6.1 µl F111 ( XbaI, PstI) + 3.9 µl F63 (SpeI, PstI) with 2 µl Ligase buffer, 7 µl ddH2O, 1 µl T4-Ligase for 40 minutes
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate).
Retrafo of P123 (digest. GSY3-BAP)
Investigator: Niklas
Procedure:
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 1 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate).
Digestion of quickchanged mRuby (without StrepTag) and transformation of E. coli XL1-Blue afterwards
Investigator: Niklas
Procedure:
- the PCR-product is digested by 1 µl DpnI (cuts methylated DNA)
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate (=rescue plate).
- next steps: 4 Minipreps, test digestion, Sequencing
QuickChange-PCR of P100 (O23 and O23)
Investigators: Julian, Niklas
Aim of the experiment: Changing of amino acid in mRuby3
Procedure:
- the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added)
- Annealing at 55°C
| volume | reagent |
| 1.25µl | O23/O24 |
| 0,5 µl | dNTP-mix |
| 1 µl | 1:100 P100 |
| 2.5 µl | Pfu Ultra II-buffer |
| 0.5 µl | Pfu Ultra II Polymerase |
| 19.25 µl | dd H2O |
| 4 x 25 µl | TOTAL |
Repeated digestion of P75 (RFP-Generator) with HincII + dephosphorylation
Investigator: Julian
Aim of experiment: Blunt-Ligation of BirA into pSB1C3
Procedure:
- Digestion of P75 (RFP-Generator) with HincII + dephosphorylation
| volume | reagent |
| 6 µl | Plasmid DNA (P75) |
| 2 µl | CutSmart buffer (10x) |
| 1 µl | HincII |
| 1 µl | Fast AP (for dephosphorylation) |
| 11 µl | ddH2O |
| =20 µl | TOTAL |
- Digestion was incubated for 2 hours at 37°C
Ligation of phosphorylated F102 (PCR of BirA) into F113 (RFP-Generator P75 digest. HincII)
Investigator: Julian
Aim of experiment: Blunt-Ligation of BirA into pSB1C3
Procedure:
- The phosphorylated DNA (prepared on July 8th) was ligated into F113 (HincII-digested RFP-Generator) as follows:
| volume | reagent |
| 4,5 µl | Vector F113 |
| 5,7 µl | Insert (BirA) |
| 2 µl | Ligase Buffer (10x) |
| 1 µl | T4 Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- next steps: Transformation into E.coli NEB Turbo.
Tuesday, July 12
Transformation of BirA–RFP generator ligation (F114) and Re-Transformation of P114
Investigator: Vivien
Aim: Transform NEB Turbo cells
Procedure:
- Followed standard transformation protocol (SOP) using 1 μL and 10 μL of P114 and F114, respectively
- Incubated at 37 °C for 1:10 h
- Plated on LB Cam (50 μL + rescue)
- Incubated at 37 °C (start 12:30)
Arabinose Induction Experiment: pBAD–GFP
Investigator: Vivien
Aim: Test different induction concentrations of arabinose
Procedure:
- Of 30 °C o/n culture, filled 5 mL each into four 10 mL culture tubes
- Induced with 0, 0.1, 1, and 10 mM final conc. of arabinose
- Took 0.5 mL sample each and stored at 4 °C
- Incubated 5 mL cultures at 30 °C for 4 h
- Took another 0.5 mL sample each and stored at 4 °C
Picking Colonies of F101 (P69 Middle Linker – P112 A3C5)
Investigator: Vivien
Aim: Pick transformed colonies
Procedure:
- Picked 3 colonies off plate with transformed cells
- Let grow at 37 °C in LB Amp (start 11:50 am)
Picking Colonies of F123 (BAP)
Investigator: Vivien
Aim: Pick transformed colonies
Procedure:
- Picked 2 colonies off plate with transformed cells
- Let grow at 37 °C in LB Cam (start 17:00)
Verification of succesful Quickchange of P141 (pASK75 with Streptactin)
Investigator: Niklas
Procedure:
- the Plasmid (5 µl) is digested by 0.5 µl NdeI (Addiition of 1 µl cut smart and 3.5 µl ddH2O)
- since the quickchange should eliminate the second Nde-restriction site, there should be one band on the gel
result:
- there is no band on the gel (the experiment was repeated multiple times showing the same results)
- therefore new primers are designed and ordered
Refolding of Streptavidin
Investigator: Niklas
Procedure:
- the sample (pelett dissolved in 6M GdmCl)was spun down (4°C, 20 mins, 18,000 rpm)
- the supernatant was transferred carefully into a falcon tube and the pellet was cast away
- the supernatant was added dropwise to 1 X PBS (500 ml) and stirred over night
Colony PCR of F110 (BAP+Nanoluc)
Investigator: Vivien, Niklas
Aim: Amplification
Procedure:
- samples were prepared with 5 µL Q5-Mastermix, 1 µl VF2-, 1 µL VR-Primer, 2 µL ddH20
- four clones were picked and added to the samples respectively
- PCR- cycles according to SOP
- Lane 1: 1 kb Ladder, Lane 2-5: clones 1-4, 10 µl each
Preparative digestion and gelelectrophoresis of P37 and P26
Investigator: Julian
Aim of experiment: Preparative digestion and gelelectrophoresis of P137 (BM40)and P26 (CMV)
Procedure:
- Batches for analytical digestions:
- P137: EcoRI and XbaI
- P26: EcoRI and SpeI
| volume | reagent |
| 23/7 µl | Plasmid DNA P137/P26 |
| 19/35 µl | ddH2O P137/P26 |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme(10 U/µl) |
| 50 µl | TOTAL |
- Preparative gelelectrophoresis was performed at 90 V for 60 min
- The whole PCR-product was applied to a 1% agorose gel. After runnig the gel two lines where visible: one at 1000Bp and one at ~500Bp.
- The 600/2100 bp band (P26/P137) was cut out and purified with the Gelextraction kit according to manufacturer's protocoll. Concentration: 20.4/33.1 ng/µl. (F115/F116)
Ligation of F115 into F116
Investigator: Julian
Aim of experiment: Ligation of F115 (CMV) into F116 (BM40 in pSB1C3)
Procedure:
| volume | reagent |
| 4,3 µl | Vector F116 (BM40 in pSB1C3) |
| 5,7 µl | Insert F115(CMV) |
| 2 µl | Ligase Buffer (10x) |
| 1 µl | T4 Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- next steps: Transformation into E.coli NEB Turbo.
Transformation of F117 (CMV in pSB1C3 with BM40), F118 (QC P100), F119 (QC P80) and P26 (CMV; Retrafo)
Investigator: Julian
Aim of the experiment: Transformation of CMV in pSB1C3 with BM40 (F117), QC P100 (F118), QC P80 (F119) and P26 in NEB Turbos(F117)/XL1 blue
Procedure: transformation in competent E. coli NEB Turbo (F117)/XL1 blue (rest) according to protocol + rescue
Result: plates (LB Cam) in incubator for further processing (37 °C)
Wednesday, July 13
QuickChange-PCR of P80 (O23 and O24)
Investigators: Christoph
Aim of the experiment: Changing of amino acid in mRuby3
Procedure:
- the batches were split, each batch contained only one primer, the batches were pooled after 15 cycles (1 µl Polymerase is added)
- Annealing at 55°C
| volume | reagent |
| 1.25µl | O23/O24 |
| 0,5 µl | dNTP-mix |
| 1 µl | 1:100 P80 |
| 2.5 µl | Pfu Ultra II-buffer |
| 0.5 µl | Pfu Ultra II Polymerase |
| 19.25 µl | dd H2O |
| 4 x 25 µl | TOTAL |
Cycling was performed according to SOP.
MiniPrep of Middle Linker-A3C5 construct
Investigator: Christoph
Aim of the experiment: MiniPrep two colonies of Middle Linker-A3C5 construct
Procedure:
- MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Both plasmids were sent to sequencing without analytical digestion because of the short lenght
Cloning of F120 (CMV-IgKappa) and F121 (CMV+EGFR / Nanoluc+StrepTag+PolyA)
Investigator: Christoph
Aim of the experiment: Ligations of F120 (CMV-IgKappa) and F121 (CMV+EGFR / Nanoluc+StrepTag+PolyA) and transformation
Procedure:
- Ligation mixtures were prepared as following:
- F120: 5 μL F63, 5 µL F111, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- F121: 3 μL F107, 7 μL F112, 2 μL Ligase buffer, 7 μL ddH2O, 1 μL Ligase
- Ligation was incubated for 30 min. at room temperature
- transformation was performed according to SOP (with 100 µl cells and 20 µl ligation mix) and plating in LB+Cam agar
Thursday, July 14
MiniPreps of Overnight Cultures
Investigator: Vivien
Aim: Prep plasmid DNA of P26, F110, F117, F114, P114, and P118
Procedure:
- MiniPrep performed according to manufacturer's instructions with Qiagen kit
- Prior to eluting, 50 μL EB buffer was applied to the membrane, and the columns were heated at 37 °C for 7 min
Results: Plasmid DNA solutions of the following concentrations: P26 (a: 55.1 ng/μL; b: 48.1 ng/μL); F110 (K2: 150.1 ng/μL; K3: 173.0 ng/μL); F117 (a: 257.7 ng/μL; b: 159.0 ng/μL; CMV/BM40: 133.2 ng/μL); F114 (83.1 ng/μL); P114 (a: 26.1 ng/μL; 170.7 ng/μL); P118 (F118?) (a: 248.0 ng/μL; 53.2 ng/μL; 241.3 ng/μL). F110 (K2: FR12904474; K3: FR12904473) and F117 (a: FR12904477; b: FR12904476; CMV/BM40: FR12904475) were sent for sequencing with VF2.
DpnI Digest of mRuby3 QC and Transformation
Investigator: Vivien
Aim: Digest original-sequence plasmid and transform obtained pure quick-changed plasmid
Procedure:
- QC was digested with DpnI (40 μL QC, 5 μL 10× CutSmart buffer, 3.5 μL ddH2O, 1.5 μL DpnI = 50 μL) at 37 °C for 2 h
- Digest (10 μL) was directly transformed into Turbo cells following SOP (including 1 h incubation time prior to plating due to chloramphenicol resistance)
- Plated 50 μL on LB Cam + rescue plate
- Incubated at 37 °C
Result: two plates; remaining digest stored at 4 °C in styrofoam tube rack
Friday, July 15
Inoculation of mRuby Quick change, F120 (CMV+IgKappa) and F121 (CMV+EGFR/Nanoluc+StrepTag+PolyA) preculture and miniprep
Investigator: Niklas, Clemens
Procedure:
- 6 colonies were picked from plate
- Cells were inoculated in 5 mL LB medium (Cam 1:1000)
- Fragments were minipreped following manufacturer's protocol (QiaPrep, Qiagen)
- Fragments were renamed receiving a P number
Quickchange of P88 (pASK75(Streptactin)) with O-029 and O-030
Investigator: Clemens
Aim: Quickchange pASK75(Streptactin) to erase one NdeI restriction site
Procedure:
- For quickchange two seperate batches were prepared:
- 1. batch: 0.7 µL O-029, 0.5 µL dNTP mix, 2.5 µL buffer, 0.5 µL Pfu ultra polymerase, 1.5 µL P88
- 2. batch: 0.7 µL O-030, 0.5 µL dNTP mix, 2.5 µL buffer, 0.5 µL Pfu ultra polymerase, 1.5 µL P88
- first stage of quickchange PCR was performed as following:
| Step | Temperature [°C] | Time [s] |
| initial Denaturation | 98 | 30 |
| 15 cycles | 98 | 10 |
| 55 | 30 | |
| 68 | 5 | |
| final Extension | 68 | 5 |
| Hold | 4 | ꝏ |
- PCR batches were mixed and 0.5 µL pfu ultra polymerase was added
- Second stage of quickchange PCR was performed as in the following:
| Step | Temperature [°C] | Time [s] |
| initial Denaturation | 98 | 30 |
| 35 cycles | 98 | 10 |
| 55 | 30 | |
| 68 | 5 | |
| final Extension | 68 | 5 |
| Hold | 4 | ꝏ |
Cloning of P79 (Nanoluc) into P147 (pSB1C3(BAP))
Investigator: Clemens
Aim:
Procedure:
- 7 µL P79 were digested with NgOMIV and PstI
- 10 µL P147 were digested with AgeI and PstI
- Digestion products were run on a 1% agarose gel
- Both fragments could be cut from the gel
- Fragments were purified by gelectraction following manufacturer's protocol (Qiagen)
- Fragments were renamed:
- digest P78= F123
- digest P147= F124
- 9,5 µL F124 and 0.5 µL F123 were ligated following manufacturers protocol (T4 DNA ligase, Thermo scientific)
- NEB Turbo were transformed with 7 µL of F125 (ligation F123+F124) following SOP
- Cells were plated on CAM LB medium and placed in the incubator at 37°C
Cloning of P145 (Middle Linker +A3C5) and P146 (Middle Linker +A3C5) into F70 (Vector fragment pSB1C3)
Investigator: Niklas, Clemens
Procedure:
- Ligation:
- 2,5 µL P145 and 1 µL P146 were pooled and digested with NgOMIV and SpeI for 1h at 37°C.
- Full digestion was ligated into 1 µL F70
- CaCl2-competent E. coli XL1-Blue cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Add 1 ml LB medium to each tube
- Incubation for 55 min at 37 °C in the 180 rpm cell-culture shaker
- 60 µl of the cell suspension was plated on one chloramphenicol plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 60 µl of LB-medium and this concentrated cell suspension was plated again on a chlorampenicol plate (=rescue plate).
Test digestion of P164 (BirA blunt end ligation) (after miniprep) and P161, P162, and P163 (all QC mRuby)
Investigator: Niklas, Clemens
Aim: Test digestion to see which plasmids contain the right insert
Procedure:
- 150 ng of plasmid DNA were digested with EcoRI and PstI following SOP
- The whole batch was applied to a 1% gel to check for cut inserts.
Results:
FOTO
- Blunt end ligation seems to not have worked (P164)
- P161, P162 and P163 all still carry the insert
Sequenced: P161= FR12904472 (NA)
Harvesting of Streptavidin
Investigator: Niklas
Aim of the experiment: Recombinant expression and purification of streptavidin
Procedure:
- After precipitation in 70 % ammonium sulfate the soluton was spun down in the centrifuge (4°C, 11000 rpm, 40 mins)
- The supernatant was discarded, and the pellet was dissolved in 2.2 M ammonium sulfate (18 mL)
- the sample was spun down in the centrifuge (4°C, 15000 rpm, 30 mins)
- the pellet was dissolved in 1X PBS(16 mL)
- next steps: last centrifugation and dialysis against Tris/Hcl (see plan on board)
Week 10, July 18–24
Monday, July 18
Inoculation of precultures for miniprep of F125 (pSB1C3(BAP-NanoLuc)) and F122 (pSB1C3(Middlelinker-A3C5))
Investigator: Clemens
Aim of the experiment:
- Miniprep, analytical digestion, analytical gel, sequencing
Procedure:
- Three colonies were picked from each plate (F125, F122)
- Inoculation was performed according to SOP
- Cells grew for 5h
- Cells onlygrew in cultures with F125 (In cultures with F122 there was no pelet)
- Miniprep was performed according to manufactrer's protocol
- Samples were renamed:
- F125 K1: P168
- F125 K2: P169
- F125 K3: P170
- Analytical digestion:
- Each plasmid was digested with EcoRI and PstI
| volume | reagent |
| 6 µl | Plasmid DNA P168/P169/P170 |
| 1 µl | Cutsmart (10x) |
| 0.5 µl | EcoRI(10 U/µl) |
| 0.5 µl | PstI (20 U/µl) |
| 1 µl | ddH2O |
| =10 µl | TOTAL |
Results:
Foto:
- Lane 1: 1kb ladder
- Lane 2: P168
- Lane 3: P169
- Lane 4: P170 --> right signals (2000 bp, ~600 bp(BAP-NanoLuc)
- Send for sequencing:
- P170: FR12904468 (VF2)
Analytical digestion of P149 , P150 (both pSB1C3(CMV-IgKappa)), P151, P152 (both pSB1C3(CMV-EGFR-NanoLuc-StrepTag-polyA)), P153, P154 (both QC pSB1C3(mRuby3))
Investigator: Clemens
Aim of the experiment: Analytical digeston to find out if the cloning was successful
Procedure:
- measure concentrations of Plasmids (see inventary list)
- Analytical digestion:
- Each plasmid was digested with EcoRI and PstI
| volume | reagent |
| 3 µl | Plasmid DNA P149/ P150/ P151/ P152/ P153/ P154 |
| 1 µl | Cutsmart (10x) |
| 0.5 µl | EcoRI(10 U/µl) |
| 0.5 µl | PstI (20 U/µl) |
| 5 µl | ddH2O |
| =10 µl | TOTAL |
Results:
Foto:
- Lane 1: 1kb ladder
- Lane 2: P149 --> right signals (2000bp; 730bp)
- Lane 3: P150 --> right signals (2000bp; 730bp)
- Lane 4: P151
- Lane 5: P152 --> right signals (2000bp; 1626bp)
- Lane 6: P153 --> very weak signal
- Lane 7: P154 --> right signals (2000bp; 700bp)
- Send for sequencing:
- P150: FR12904471 (VF2)
- P152: FR12904470 (VF2)
- P154: FR12904469 (VF2)
Transformation of QC of P88 (pASK75(Streptactin))
Investigator: Clemens
Aim of the experiment:
- See if quickchange has worked
Procedure:
- NEB Turbo cells were transformed with 10 µl of QC P88 according to SOP
- Cells were plated on LB medium (Amp) and incubated at 37°C
Digestion of P165 (pSB1C3(CMV-BM40) and cloning of F112 (Nanoluc-StrepTag-polyA) into P165
Investigator: Clemens
Aim of the experiment:
- Create biobrick: CMV-BM40-Nanoluc-StrepTag-polyA
Procedure:
- Preparative digestion batch was prepared as in the following:
| volume | reagent |
| 15 µl | Plasmid DNA P156 |
| 3 µl | Cutsmart (10x) |
| 1 µl | AgeI(10 U/µl) |
| 1 µl | PstI(20 U/µl) |
| 10 µl | ddH2O |
| =30 µl | TOTAL |
- Digestion was seperated by gelelectrophoresis
Results:
Foto:
- Lane 1: 1kb ladder
- Lane 2: F126 (digest P165) --> right length
Procedure:
- Fragment was cut from the gel and purified by gelextraction which was performed by manufacturer's protocol (Qiagen)
- F126 was ligated with F112
Ligation batch:
| volume | reagent |
| 2 µl | Vector DNA F126 |
| 18 µl | Insert DNA F112 |
| 3 µl | T4 ligation buffer (10x) |
| 6 µl | T4 ligase |
| µl | ddH2O |
| =30 µl | TOTAL |
- Ligation was incubated at 37°C for 30 min
- NEB Tubros were transformed with F127 (lig. F112+F126)
- Cells were plated on LB medium (Cam)
Tuesday, July 19
Inoculation of Precultures of F127 (pSB1C3(CMV-BM40-Nanoluc-StrepTag-polyA))
Investigator: Clemens
Aim: Prepare cells for miniprep of plasmids
Procedure:
- six colonies were picked and inoculated in LB medium (CAM)
- Plates were incubated on a shaker at 37°C
Results:
- cellgrowth ?
Reamplifiying Streptactin from GSY6
Investigator: Christoph
Aim: Reamplifiying Streptactin from GSY6 for further processing in QC reaction (delete NdeI site from pASK)
Procedure:
- PCR reaction was prepared according to SOP
- Cycling steps were taken from the QC protocol because no further cycler was free
Results:
- will be available after DpnI digestion and transformation
QuickChange-PCR of pASK75 with O29 and O30
Investigator: Christoph
Aim: Deleting NdeI from pASK (P10)
Procedure:
- QC-PCR was performed according to SOP
- Primers: O-029 and O-030
- Annealing at 55°C
- Elongation time: 5 min
- 2-step protocol was applied
Miniprep of E. coli Xl1-Blue P171, P172 and P173 (pSB1C3(CMV-BM40-Nanoluc-StrepTag-polyA))
Investigator: Javier
Aim of the experiment: Extracting P171, P172 and P173 (CMV-BM40-Nanoluc-StrepTag-polyA) from E.coli XL-1-blue
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations:
| Plasmid | c [ng/µl] |
| P171 | 75,4 |
| P172 | 108,2 |
| P173 | 66,4 |
Transformation of F136 (A3C5+Middle-Linker), F137 (EGFR+mRuby-Strep), F138 (mRuby3-Strep+PolyA) and P161 (QC mRuby-Strep)
Investigator: Javier
Aim of the experiment: Transformation of A3C5+Middle-Linker (F136), EGFR+mRuby-Strep (F137), mRuby3-Strep+PolyA (F138) and QC mRuby-Strep (P161) XL1 blue
Procedure: transformation in competent E. coli XL1 blue according to protocol + rescue
Result: plates (LB Cam) in incubator for further processing (37 °C)
Colony PCR of F127 (CMV-BM40-Nanoluc-StrepTag-polyA)
Investigator: Clemens
Aim: See if cloning was successfull
Procedure:
- samples wwere prepared with 5 µL Q5-Mastermix, 1 µl VF2-, 1 µL VR-Primer, 2 µL ddH20
- six clones were picked and added to the samples respectively
- PCR- cycles according to SOP (Annealing-temp: 66°C; Ampflification time: 1.5min)
Results:
- Lane 1: 1 kb Ladder
- Lane 2: F127 K1
- Lane 3: F127 K2
- Lane 4: F127 K3
- Lane 5: F127 K4
- Lane 6: F127 K5
- Lane 7: F127 K6
Preparative digestion of P161 (pSB1C3(mRuby3-StrepTag)), P73 (pSB1C3((PolyA)), P96 ((pSB1C3(EGFR-TMD)), P69 (pSB1C3(Middle-linker)) and P112 (pSB1C3(A3C5))
Investigator: Clemens, Christoph, Javier
Aim: Create biobricks
Procedure:
- Preparative digestion of P161 and P73
| volume | reagent |
| 15 µl/ 20 µl | Plasmid DNA P161/ P73 |
| 3 µl/ 3 µl | Cutsmart (10x) |
| 1 µl/ 1 µl | EcoRI/ EcoRI |
| 1 µl/ 1 µl | SpeI/ XbaI |
| 10 µl/ 5 µl | ddH2O/ ddH2O |
| =30 µl | TOTAL |
- Preparative digestion of P96 and P161
| volume | reagent |
| 10 µl/ 15 µl | Plasmid DNA P96/ P161 |
| 3 µl/ 3 µl | Cutsmart (10x) |
| 1 µl/ 1 µl | PstI/ PstI |
| 1 µl/ 1 µl | AgeI/ NgOMIV |
| 15 µl/ 10 µl | ddH2O/ ddH2O |
| =30 µl | TOTAL |
- Preparative digestion of P69 and P112
| volume | reagent |
| 20 µl/ 15 µl | Plasmid DNA P69/ P112 |
| 3 µl/ 3 µl | Cutsmart (10x) |
| 1 µl/ 1 µl | EcoRI/ EcoRI |
| 1 µl/ 1 µl | AgeI/ NgOMIV |
| 5 µl/ 10 µl | ddH2O/ ddH2O |
| =30 µl | TOTAL |
- Digestions were incubated at 37°C for 2h
- Samples were applied to a 1% agarose gel (40 min; 90V)
- Samples were cut from the gel afterwards:
- Renamed:
- digest P69: F128
- digest P112: F129
- digest P161: F130
- digest P96: F131
- digest P161: F132
- digest P73: F133
Results:
Fotos
Procedure:
- Gelextraction was performed after manufacturers protocol (Qiagen)
- After gelextraction concentrations were measured:
- Ligation:
| volume | reagent |
| xx µl | Vektor F129 |
| xx µl | Insert F128 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| x µl | ddH2O |
| =20 µl | TOTAL |
| volume | reagent |
| xx µl | Vektor F131 |
| 7xx µl | Insert F130 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| x µl | ddH2O |
| =20 µl | TOTAL |
| volume | reagent |
| xx µl | Vektor F133 |
| 7xx µl | Insert F132 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| x µl | ddH2O |
| =20 µl | TOTAL |
- Ligation was performed according to SOP
- Xl1 blue cells were transformed with the Ligations according to SOP
Wednesday, July 20
Analytical digestion and gelelectrophoresis of P171-P173 & QC of P10
Investigator: JB, MT
Aim of experiment: Check if everything is fine.
Procedure:
- Digest P171-P173 with XbaI + SpeI and QC of P10 with NedI and incubate 1 h at 37 °C
- Run 1 % agarose-gel at least 40 min at 100 V
Results:
Lane1: 1Kb ladder; Lane2: P171; Lane3: P172; Lane4: P173; Lane5: QC of P10
=> P171 was sent for sequencing: FR12904466 => NdeI test digestion shows two signals although there should have been just one
Preparation of GSY 6; F134; F135 for ligation
Investigator: JB, MT
Aim of experiment: Prepare GSY6 for Ligation.
Procedure:
- Digestion of GSY6 with SpeI + HindIII
- Preparative 1 % Agarose gel 100 V for at least 40 min.
- Gel extraction of GSY6, F134, F135 by Quiagen kit.
Results:
- GSY6 -> F139 : 2.5 ng/µL
- F134 : 4 ng/µL
- F135 : 10.2 ng/µL
Preparative agarose gel GSY6:
Ligation & transformation of F134 + F63; F135 + F139
Investigator: JB, MT
Aim of experiment: Ligation & transformation of F134 + F63; F135 + F139
Procedure:
Ligation following SOP(13.02.2013)
- F134 + F63 -> F140
- F135 + F139 -> F141
transformation following SOP(13.02.2013)
- F140 on CAM
- F141 on AMP
Results:
We will see tomorrow, please update this section.
Thursday, July 21
Miniprep of F136 (K1 & K2) (pSB1C3(Middle linker-A3C5)), F137 (K1 & K2) (pSB1C3(EGFR TMD-mRuby3-StepTag)), F138 (K1 & K2) (pSB1C3(mRuby3-StrepTag-polyA) and retrafo of P161 (pSB1C3(mRuby3-StrepTag))
Investigator: Clemens
Aim of experiment: Get plasmids for analytical digestion and sequencing
Procedure:
- Minipreps were performed according to manufacturers protocol (Qiagen, QiaPrep)
- Samples were renamed afterwards:
- F136 K1: P174
- F136 K2: P175
- F137 K1: P176
- F137 K2: P177
- F138 K1: P178
- F138 K2: P179
- retrafo of P161: P180
- Analytical digestion:
| volume | reagent |
| 1 µl | Plasmid DNA |
| 1 µl | Cutsmart (10x) |
| 0.5 µl | EcoRI |
| 0.5 µl | PstI |
| 7 µl | ddH2O |
| =10 µl | TOTAL |
- Ligation was renamed: F143
- Digestion products were seperated via gelelectrophoresis
Results:
Foto:
- All plasmids seemed to carry the appropriate insert
Sequencing:
- P175: FR12904465 (VF2)
- P177: FR12904464 (VF2)
- P178: FR12904463 (VF2)
Preparative digestion of P116 (pSB1C3(IgKappa))
Investigator: Clemens, Christoph
Aim of experiment: cloning of CMV in front of IgKappa signal peptide
Procedure:
- preparative digestion batch
| volume | reagent |
| 10 µl | Plasmid DNA P116 |
| 3 µl | Cutsmart (10x) |
| 1 µl | EcoRI |
| 1 µl | XbaI |
| 15 µl | ddH2O |
| =30 µl | TOTAL |
- Digestion was applied to a 1% agarose gel (45 min; 90V)
- Sample was cut from the gel and gelextraction was performed according to manufacturers protocol
- Ligation of Fx and F115 (digest CMV promotor):
- Ligation:
| volume | reagent |
| 3.9 µl | Vektor F142 |
| 6.1 µl | Insert F115 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 20 µl | ddH2O |
| =20 µl | TOTAL |
- Transformation of XL1blue was performed according to SOP
- Cells were plated on LB medium (CAM) and incubated over night at 37°C
Friday, July 22
Analytical digestion of pASK_T7 RBS_LCN2_His
Investigator: Luisa
Aim of the experiment: Analytical digestion to find out whether there is more than one NdeI restriction site.
Procedure:
- Analytical digestion:
- The plasmid was digested with NdeI and HindIII
| volume | reagent |
| 3 µl | Plasmid DNA |
| 1 µl | Cutsmart (10x) |
| 0.5 µl | NdeI |
| 0.5 µl | HindIII |
| 5 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- Two bands were visible what means that there is only one NdeI restriction site.
Inoculation of Precultures of F143 (pSB1C3(CMV+IgKappa))
Investigator: Luisa
Aim: Prepare cells for miniprep of plasmids
Procedure:
- two colonies were picked and inoculated in LB medium (CAM)
- Plates were incubated on a shaker at 37°C over night.
Harvesting of Streptavidin
Investigator: Niklas
Aim of the experiment: Recombinant expression and purification of streptavidin
Procedure:
- the solution was spun down in the centrifuge (4°C, 15000 rpm, 20 mins) to remove last contamination
- The pellet was discarded and the supernantant was dialysed against 20 mM Tris/HCl (ph 8) witt 50 mM NaCl
- last step to do: steril filtration
- the concentration was determined via UV/VIS: the absorption at 280 nm is 0.469 (1:100 dilution), this equals a concentration of 62 mg/mL (about 16 mL) (mass 54 kDa; Absorbtion coefficient 40455 1/M*cm
Sequencing of P174(Middle Linker + A3C5)
Investigator: Niklas
Procedure:
- Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Cloning of P178 (mRubyStrepPolyA) / P180 (mRubyStrep) behind EGFR-TMD in pSB1C3 (P96)
Investigator: Niklas
Procedure:
- Preparative digestion:
- The Inserts were digested with PstI and NgoMIV
| volume | reagent |
| 7.7 / 8.2 µl (about 3µg) | Plasmid DNA |
| 2 µl | Cutsmart (10x) |
| 1 µl | PstI |
| 1 µl | NgoMIV |
| 8.3 / 7.8 µl | ddH2O |
| =20 µl | TOTAL |
- both fragments were ligated with F131 (digest P96 [AgeI; PstI])
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
- Results: Cloning didn't work. No colonies on plate. Repeat.
Cloning of Middle Linker before A3C5 in pSB1C3 (Repeat)
Investigator: Julian, Niklas
Procedure:
- Preparative digestion:
- The Middle Linker (P69) was digested with EcoRI and AgeI
| volume | reagent |
| 30 µl | Plasmid DNA |
| 5 µl | Cutsmart (10x) |
| 1.5 µl | EcoRI |
| 1.5 µl | AgeI |
| 12 µl | ddH2O |
| =50 µl | TOTAL |
- the fragment was ligated with F129(digest P112 [EcoRI; NgOMIV])
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
Week 11, July 25th - July 31st
Monday, July 25
Inoculation of Precultures of F143 (pSB1C3(CMV+IgKappa)), F141new (pASK(Streptactin)), F151 (pSB1C3(middleLinker+A3C5), pASK75_T7-RBS-LCN2-His, F148 (pSB1C3(mRubyStrepPolyA+ CMV-EGFR Signal-Pept)
Investigator: Luisa
Aim: Prepare cells for miniprep of plasmids
Procedure:
- Three colonies of each were picked and inoculated in LB medium (CAM) respectively (AMP) fpr pASK.
- Plates were incubated on a shaker at 37°C for 6 hours.
MiniPreps and analytical digestion of F143 cultures
Investigator: Luisa
Aim: Prep plasmid DNA of F143 (pSB1C3(CMV + IgKappa)) and checking wheather ligation was successful.
Procedure:
- MiniPrep performed according to manufacturer's instructions with Qiagen kit
- Prior to eluting, 30 μL EB buffer was applied to the membrane since the pellet was quite small, and the columns were heated at 37 °C for 7 min
Results:
- Concentrations: K1 = 230,4ng/µl, K2 = 162,4 ng/µl
Analytical Digestion:
- An 20µl batch was prepared for analytical digestion for each of the two clones:
| volume | reagent |
| 10 µl | Plasmid DNA |
| 2 µl | Cutsmart (10x) |
| 1 µl | EcoI |
| 1 µl | PstI |
| 6 µl | ddH2O |
| =20 µl | TOTAL |
Results:
- Two bands were visible at 750 Bp which is the size of IgKappa and CMV together. Cloning was successful. Sequence still has to be ckecked though.
Cloning of P178 (mRubyStrepPolyA) / P180 (mRubyStrep) behind EGFR-TMD in pSB1C3 (P96). Repetition.
Investigator: Luisa
Aim: See title.
Procedure:
- Digestion of P178 and P180: 7µg of DNA were digested.
| volume | reagent |
| 18 µl | Plasmid DNA |
| 5 µl | Cutsmart (10x) |
| 2,5 µl | NgoMIV |
| 2,5 µl | PstI |
| 22 µl | ddH2O |
| =50 µl | TOTAL |
- Digestion at 37°C for 2,5h.
Cloning of NanoLuc-Strep-PolyA (F112) behind CMV-IgKappa in pSB1C3 (P181).
Investigator: Luisa
Aim: See title.
Procedure:
- Digestion of P181:
| volume | reagent |
| 10 µl | Plasmid DNA |
| 5 µl | Cutsmart (10x) |
| 2,5 µl | AgeI (20u/µl) |
| 2,5 µl | PstI (20u/µl) |
| 30 µl | ddH2O |
| =50 µl | TOTAL |
- Digestion at 37°C for 2,5h.
Digestion and gel-extraction of P178; P180
Investigator: MT
Aim of experiment: Preparation of P178 and P180 for ligation.
Procedure:
- Digest P178; P180 with NgoMIV + PstI incubate 1 h at 37 °C
- Run 1 % agarose-gel at least 40 min at 100 V
- Gel extraction by Quiagen kit.
Results:
- P178 [NgoMIV +PstI digest] -> F152: 17 ng/µL
- P180 [NgoMIV +PstI digest] -> F153: 8.1 ng/µL
Ligation & transformation of F131 + F152; F131 + F153
Investigator: MT
Aim of experiment: Ligation & transformation of F131 + F152; F131 + F153
Procedure:
Ligation following SOP(13.02.2013)
- F131 + F152 -> F154
- F131 + F153 -> F155
Transformation following SOP(13.02.2013)
- F154 E. coli. turbo CAM
- F155 E. coli. turbo CAM
Results:
We will see tomorrow, please update this section.
Wednesday, July 27th
Minipreps of EGFR-TMD - mRuby - Strep - PolyA (F131+F152) and EGFR-TMD - mRuby - Strep (F131+F153)
Investigator: JB
Results: Procedure according to protocol (spin kit). Resulting plasmids:
EGFR-TMD - mRuby - Strep - PolyA (F131+F152) -> P196, P197 EGFR-TMD - mRuby - Strep (F131+F153) -> P198, P199
Concentrations: P196: 228 ng/µl P197: 212 ng/µl P198: 197 ng/µl P199: 322 ng/µl
Thursday, July 28th
Inoculation of Precultures of F141new (pASK(Streptactin))
Investigator: Luisa
Aim: Prepare cells for miniprep of plasmids
Procedure:
- Three colonies of each were picked and inoculated in LB medium (CAM) respectively (AMP) fpr pASK.
- Plates were incubated on a shaker at 37°C for 6 hours.
PCR of GSY6
Investigator: Luisa
Aim: Amplification of Streptavidin mutant and Traptactin for further cloning into pASK.
Procedure:
- reaction was performed after SOP (PCR Using Q5® High-Fidelity DNA Polymerase, NEB)
- 2ng Plasmid were used for amplification (1 µL)
- Primers: VR and VF2
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial Denaturation | 98 | 30 |
| 35 cycles | 98 | 10 |
| 66 | 30 | |
| 72 | 90 | |
| final Extension | 72 | 5 |
| Hold | 4 | ꝏ |
- PCR product was purified accoding to manufacturer's protocol (PCR puification kit, Qiagen)
- DNA was eluted 40 µL EB buffer
Digestion of pASK
Investigator: Luisa
Aim: Preparing pASK for ligation with Streptavidin mutant and Traptactin.
Procedure:
| volume | reagent |
| 20 µl | Plasmid DNA |
| 5 µl | Cutsmart (10x) |
| 1,5 µl | NdeI |
| 1,5 µl | HindIII |
| 22 µl | ddH2O |
| =50 µl | TOTAL |
Cloning of Traptactin and Streptavidin mutant into pASK (P191).
Investigator: Luisa
Aim: Preperation of the plasmids for Streptavidin and Traptactin expression in E.coli.
Procedure:
- Cutting out the crucial bands from the gel containing the PCR-product and the digested plasmid (P190) and purifiing them with the Gelextraction Kit according to the protocoll. First Gel failed: Very small band with wrong length...--> Repetition of digestion with P190.
- The PCR product of GSY6 was extracted from the gel (Band at ~800 Bp) with the Gelextraction Kit and then digested with NdeI and HindIII to get the fragments Streptavidin m1 and Traptactin. Inactivation was performed at 80°C for 10 min.
- Concentrations: 33,9ng/µl (GSY6),
- Ligation...
Cloning of NanoLuc-Strep-PolyA (F112) behind CMV-IgKappa in pSB1C3 (P182). Repetition.
Investigator: JB
Aim: See title.
Procedure:
- Digestion of P182:
| volume | reagent |
| 15 µl | Plasmid DNA |
| 3 µl | Cutsmart (10x) |
| 1,5 µl | AgeI (20u/µl) |
| 1,5 µl | PstI (20u/µl) |
| 9 µl | ddH2O |
| =30 µl | TOTAL |
- Digestion at 37°C for 2.5h.
- Take care: P182 is not sequenced
Retrafos of P178 (mRuby-Strep-PolyA) and P181 (CMV + IgKappa)
Investigator: JB
Results: Transformation according to protocol, plates were incubated at 37°C overnight.
Dialysis of eGFP
Investigator: Julian
Aim of the experiment: Preparation of eGFP for following biotinylation
Procedure:
- eGFP was poured in a dialysis hose (cut-off 14 kDa)
- The hose was then placed in 4 L ice cold borate buffer 50 mM pH 8.85 (no salt)
- The dialysis took place at 4°C over night
Sequencing of P160 (NanoLuc+StrepTag+Poly-A)
Investigator: Julian
Procedure:
- Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Friday, July 29th
Minipreps of EGFR-TMD - mRuby - Strep - PolyA (F131+F152) and EGFR-TMD - mRuby - Strep (F131+F153)
Investigator: Julian
Procedure:
Procedure according to protocol (spin kit). Resulting plasmids:
CMV+EGFR-SigPep+mRuby3+StrepT+PolyA (F131+F152) -> P200, P201 pASK_T7 RBS_LCN2_His (Vorlage für gewünschte Inserts) -> P202, P203
Results:
Concentrations: P196: 385.3 ng/µl P197: 462.6 ng/µl P198: 84.8 ng/µl P199: 96.0 ng/µl
- Send for sequencing:
- P201: FR12904454 (VF2)
Analytical digestion of P202 and P203 (pASK_T7 RBS_LCN2_His)
Investigator: Julian
Aim of the experiment: Controll whether there is more than one NdeI restriction site.
Procedure:
- Analytical digestion:
- The plasmid was digested with NdeI and HindIII
| volume | reagent |
| 2 µl | Plasmid DNA |
| 1 µl | CutSmart buffer |
| 0.5 µl | NdeI |
| 0.5 µl | HindIII |
| 6 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- Two bands were visible what means that there is only one NdeI restriction site.
Chemical biotinylation of BSA and eGFP
Investigator: Julian
Procedure:
1 g BSA was chemically biotinylated with a 40x molar excess of biotin-NHS-ester:
- 100 ml of 50 mM borate buffer with 50 mM NaCl pH 8.85
- dissolve BSA (10 mg/ml)
- add adequate amount of biotin-NHS-ester: 205 mg
- reaction for approx. 45 minutes at RT after full dissolving of biotin-NHS-ester
- store at 4 °C
eGFP was chemically biotinylated with a 16x (error in calculations) molar excess of biotin-NHS-ester:
- eGFP in 75 ml of 50 mM borate buffer without NaCl pH 8.85
- 1to10 dilution: extinction(280) = 0.371
- add adequate amount of biotin-NHS-ester: 69 mg
- reaction for approx. 45 minutes at RT after full dissolving of biotin-NHS-ester
- store at 4 °C
Next Steps
- add another 104 mg of biotin-NHS-ester incl. reaction at RT tp reach desired 40X molar excess
- Purification via column chromatography
Week 12, August 1st - August 7th
Monday, August 01
Cloning of Streptavidin/Streptactin/Traptactin from GSY5 and GSY6 into pASK75 + Transformation
Investigator: JB
- PCR products from GSY5 and GSY6 were, after digestion overnight with Nde1, loaded on an 1.8% agarose gel. The fragment bands at roughly 350 bps were extracted.
- Digested pASK75 (P202 and P203 dig. with Nde1) were loaded onto an 1% agarose gel, the vector fragments at 3800 bp isolated and purified.
- Vector fragment plus Streptavidin fragments were ligated and transformed into XL-1 blue
-> F159+F160, F159+F161
Cloning of EGFR-TMD - mRuby - Strep with IRES-BirA
Investigator: JB
- 2 µg of P77 (EcoRI, XbaI) and P199 (EcoRI, SpeI) were digested for 2 hours at 37°C
- Digestions were separated on a 1% agarose gel, fragments corresponding to the vector (P77) and to the to be cloned insert (P199) were isolated and purified
- Ligation of inserts for 30 mins at RT
- Transformation of ligation into XL1-blue
Retrafos of cell culture constructs for signal peptide testing for midipreps
Investigator: JB
- Retransformation of all constructs in XL1-blue
- Constructs concerned are: P152, P171, P205 (Nanoluc-constructs) and P201, P208 and P211 (mRuby-constructs)
- Transformation according to protocol, plates incubated at 37°C overnight
Tuesday, August 02
Wednesday, August 03
Verification of cloning P210 (CMV-IgKappa-Nanoluc-StrepTag-PolyA)
Investigator: Niklas
Aim of the experiment: Verification of successful cloning after sequencing showed that the last part of the construct might be missing
Procedure:
- digestion with AgeI and PstI (sample was digested with EcoRI and PstI for 10 minutes, than inactivated an ligated)
Results:
- the gel shows a band at about 3500 bp (pSB1C3 + full insert)
- gel shows additionally a band below 750 bp (CMV+ IgKappa)
- the sample contains the full insert as well as the small insert (CMV+IgKappa without Nanoluc-StrepTag-PolyA)
- cloning not succesful
- the construct was inocculated from plate again
Transformation of E. coli XL1 blue with Ligations F159 + F160 and F159 + F161 (pASK + streptavidin from GSY 5 and 6)
Investigator: Niklas
Aim of the experiment: Transformation of E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator.
Thursday, August 04
MidiPrep P152 and P171 (pSB1C3)
Investigator: EF
Aim of the experiment: isolate Plasmids
Procedure:
- MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen). After washing the DNA with 70% Ethanol the pellet was left in special centrifugation tubes (labeled 152+QF; 171+QF) in order to air-dry. EB buffer for storage was later added by CG.
- Conc: 1000 ng/ul
Cloning of BAP Nanoluc (P206) before A3C5 (P213)
Investigator: NA
Procedure:
- Preparative digestion:
P213 (A3C5): EcoRI and NgoMIV -> F170
P206 (BAP Nanoluc): EcoRI and AgeI -> F171
Both fragments were ligated after gelextraction.
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of Ligation was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
the plates showed no colonies. repeat
Friday, August 05
Verification of successful cloning of all Streptavidin variants into pASK75
Investigator: CG
Aim of the experiment: Minipreps von pASK75 Streptavidin variants (codon optimzied Streptavidin, Streptactin, Traptactin)
Procedure:
- MiniPrep was performed according to the manufacturer's protocol (QIAprep MiniPrep, Qiagen) for 3 clones of GSY5 and 6 clones of GSY6
- Analytic digestion with 0.25 µL NdeI, 0.25 µL HindII, 1 µL SmartCut Buffer, 200 ng plasmid DNA, H2O to 20 µl
- 10 µL on 1% agarose gel electrophoresis
Results: Successful cloning was verified for all plasmids by analytical digestion. Expected and observed fragment size for the vector was 3.1 kb and about 350 bp for the inserts. 4 plasmids were sent for sequencing (1x GSY5, 3x GSY6). Alle sequences were error-free. Therefore all 3 Streptavidin variants are available now.
Transformation of: Streptavidin variants, PAS-Plasmid, Interlab-Plasmids, Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA and Ligation of BAP-Nanoluc-A3C5
Investigator: CG
Aim of the experiment: Transformation of Streptavidin variants, PAS-Plasmid, Interlab-Plasmids into E. coli BL21 and Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA, Ligation of BAP-Nanoluc-A3C5 in E. coli XL1 blue
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of Ligations mixes were added to 50 µl of competent cells and gently mixed, 1 µl for ccc Plasmids
- 30 min incubation on ice
- 1 min. heat shock at 42 °C
- Adding of 950 µl LB-medium to each tube, and incubation for 1 h at 37°C
- The cell suspension was plated on ampicillin plates (inclusive rescue plate) / CamR- plates and incubated for 7 h at 37 °C in the programmed incubator.
Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA and Ligation of BAP-Nanoluc-A3C5
Investigator: CG
Aim of the experiment: Re-Ligation of EGFR-TMD-mRuby-Stre-IRES-BirA, Re-Ligation of BAP-Nanoluc-A3C5
Procedure:
The corresponding Fragments were mixed with 2 µl Ligase-buffer, 1 µl T4-Ligase and filled with water up to 20 µl
- 7.3 µl F163 + 2.7 µl F162 (=F173)
- 3.7 µl F170 + 6.3 µl F171 (=F174)
The Ligation mixes were incubated for 1 h at room temperature.
SDS Page of Streptavidin production
Investigator: CG
Aim of the experiment: Verification of successful Streptavidin production and preperation for the data package
Procedure: Streptavidin samples from the production process were further processed with SDS buffer. Some samples had very few volume, therefore the overall volume was reduced. For informations on the samples content please refer to NA. The low volume might be a problem due to evaporation issues.
- Sample 1: the tube was empty
- Sample 2: 1 µl sample was mixed with 7 µl water and 2 µl SDS buffer
- Sample 3: 80 µl sample was mixed with 20 µl SDS buffer
- Sample 4: 80 µl sample was mixed with 20 µl SDS buffer
- Sample 5: 20 µl sample was mixed with 5 µl SDS buffer
- Sample 6: 80 µl sample was mixed with 20 µl SDS buffer
- Sample 7: 80 µl sample was mixed with 20 µl SDS buffer
- Sample 8: 80 µl sample was mixed with 20 µl SDS buffer
- Sample 9: 80 µl sample was mixed with 20 µl SDS buffer
10 µl of each sample were loaded on the pre-mixed SDS gel.
Gel 1: 8 µl Ladder, samples 1-6
Gel 2: 8 µl Ladder, samples 7-9, 5 µl of samples 7-9
The gel ran at 160 V for about 1 1/2 h. Afterwards it was incubated for 30 min. in staining solution, 10 min. in destaining solution 1 and over the weekend in destaining solution 2.
Results:
Week 13, August 8th - August 14th
Monday, August 08
PCR, Purification and digestion of BirA
Investigator: LK
Aim of the experiment: Amplification of BirA
Procedure:
- PCR according to the SOP (primers: O17 and O18, Annealing at 62°C)
- PCR purification via the purification kit according to the manufacturer's protocol
- preparative digestion with XbaI and PstI over night in a 50µl batch
Results:'
- Concentration after PCR-purification: 116ng/µl
- Gel: One thick band at ~1000Bp --> BirA and one at ~2100Bp --> vector, we're cut out and purified via the Gelextraction kit according to the manufacturer's protocoll.
- Concentrations: BirA (F186) : 62,5 ng/µl, pSB1C3 (F187): 32,5 ng/µl
Determination of Streptavidin concentration
Investigator: NA
Procedure:
- OD280 measurement with quartz cuvette
result: the 1:100 dilution showed an OD of 0,469
Calculation with extinction coefficient (expasy tool) of 41940 M -1 * cm -1 and 13.3 kDA as molar mass of streptavidin monomer provides concentration below:
14.87 mg/mL
Inocculation of Streptavidin (wild type, P235) in M9 medium for fermentation
Investigator: NA
Procedure:
- M9 medium with Amp (1:1000) is inocculated with preculture (~50 mL) of P235
- the culture is incubated in the shaker over night at 37°C
PCR purification
Investigator: EF
Aim of the experiment: Purificate bands for blunt ligation in F113
Seeding of HEK293T cells in cell culture
Investigator: JB
Aim of experiment:
Thawing and seeeding of HEK293T cells for experiments later this week
Procedure:
- Tube with cells was thawed quickly in a 37°C waterbath
- After bringing most of the cell suspension in a fluid state, the suspension was transferred into a 50 ml Falcon tube and suspended in 50 ml of RPMI1640 medium (with 10% CFS and Pen/Strep added).
- Cells were spun down for 2 mins at 2,300 rcf and the pellet resuspended in 20 ml of medium in order to remove residual DSMO from the cells (DSMO having been used for freezing the cells).
- The resuspended cells were transferred into a culture flask and incubated overnight at 37°C/5% CO².
Cloning of IgKappa signale peptide, PAS-Lysin and further constructs for the receptor
Investigator: CG
Aim of experiment:
Cloning of CMV-IgKappa+Nanoluc-Strep-PolyA, BAP-Nanoluc+A3C5, IRES-BirA + polyA and Sumo-PAS-Lysin into pASK75
Procedure:
Digestion of 3 µg of each plasmid in 30 µl mix:
- 20 µl P205 (CMV-IgKappa) with EcoRI, AgeI -> Gel 1, left lane, expected fragment at about 600 bp -> F175
- 15 µl P160 (Nanoluc-Strep-PolyA) with EcoRI, NgoMiV -> Gel 1, right lane, expected fragment at about 3000 bp -> F176
- 20 µl P212 (A3C5) with EcoRI, NgoMiV -> Gel 2, left lane, expected fragment at about 2.200 bp -> F177
- 10 µl P206 (BAP-Nanoluc) with EcoRI, AgeI -> Gel 2, right lane, expected fragment at about 500 bp -> F178
- 10 µl P199 (EGFR-TMD-mRuby-Strep) with EcoRI, SpeI -> Gel 3, left lane, expected fragment at about 1.500 bp -> F179
- 20 µl P236 (PAS-Lysin-Sumo) with NdeI, HindIII -> Gel 3, right lane, expected fragment at about 1.000 bp -> F180
- 10 µl P77 (IRES-BirA-KDEL) with EcoRI, SpeI -> Gel 4, left lane, expected fragment at about 1000 bp -> F181
Ligation in 20 µl mix (s. above fragment names):
- F182: 2.9 µl F176 + 7.1 µl F175
- F183: 5.2 F177 + 4.8. F178
- F184: 7.0 µl F159 + 3.0 µl F180
- F185: 2.9 µl F133 + 7.1 µl F181
Transformation according to SOP.
Results:
For all ligations colonies grew the next day.
Gel 1:
Gel 2:
Gel 3:
Gel 4:
Tuesday, August 09
Transformation of Biobricks in NEB Turbo
Investigator: Julian
Aim of the experiment: Acquisition of pBAD/AraC and Lox site
Procedure:
- resuspend DNA with 10 µl ddH2O in well of interest (standard distribution kit)
- incubate for 5 min
- transformation of 1 µl plasmid according to SOP in NEB Turbo
Used bricks: BBa_I0500 and BBa_J61046
Inoculation and induction of AraC+pBAD controlled eGFP production in E.coli BL-21
Investigator: LK
Aim of the experiment: Confirmation of arabinose dependend activation of the pBAD-promotor in AraC-Operon.
Procedure:
- E.coli Bl-21 with P61 (PC+AraC+pBAD+eGFP in pSB1C3) we're picked and a 4ml preculture (LB+cam) was inoculated and incubated for 6h.
- According to the following table 50ml cultures (LB+cam) we're incoculated with 500µl of the preculture and incubated over night at 37°C.
| Arabinose concentration: | negative control | 100mM | 10mM | 1mM | 100µM | 10µM |
Results:
Cloning of F186 (BirA) into F187 (pSB1C3)
Investigator: LK
Aim of the experiment: Getting the BirA for the prokaryotic receptor.
Procedure:
- 1,7µl of F187 and 6,3µl of F186 we're ligated for 1h at RT accoridng to the SOP. --> F189
- The ligation-batch was transformed into NEB-Turbo cells and plated out on cam-plates. Incubation at 37° o/n.
Inoculation of precultures from Ligations F182-F185
Investigator: LK
Aim of the experiment: Getting plamids of CMV-IgKappa+Nanoluc-Strep-PolyA (F182), BAP-Nanoluc+A3C5 (F183),Sumo-PAS-Lysin (F184) and IRES-BirA + polyA (F185)
Procedure:
- Inoculation of 4mL cultures (LB cam resp. amp (F184)). Incubation at 37°C o/n.
Inocculation of fermenter
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- centrifuge preculture (P235 in M9 medium)
- resuspend pellet in HCDF medium and inocculate fermenter (HCDF medium)
- pO2= 60 %; Temp= 30°C, pH~7
- growth over night
SDS Page of Streptavidin production
Investigator: NA
Aim of the experiment: Verification of successful Streptavidin production and preperation for the data package
Procedure: Streptavidin samples from the production process were further processed with SDS buffer. Some samples had very few volume, therefore the overall volume was reduced.
- 1: 8 µl Ladder
- 2: 10 µl after induction (1 µl sample was mixed with 7 µl water and 2 µl SDS buffer)
- 3: 5 µl supernantant after denaturation (20 µl sample was mixed with 20 µl H2O and 10 µl SDS buffer)
- 4: 5 µl before precipitation(80 µl sample was mixed with 20 µl SDS buffer)
- 5: 10 µl supernantant after 40% precipitation (80 µl sample was mixed with 20 µl SDS buffer)
- 6: 10 µl supernantant after 70% precipitation (80 µl sample was mixed with 20 µl SDS buffer)
- 7: 10 µl after dissolving in 2.2M (NH4)2SO4 (80 µl sample was mixed with 20 µl SDS buffer)
The gel ran at 145 V
Results:
Transformation of F192 and P238
Investigator: NA
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 50 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- The cell suspensions were plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C.
Wednesday, August 10
Minipreps and analytical digestion of P237 - P243
Investigator: LK
Aim of the experiment: Isolating and verifiing plamids of CMV-IgKappa+Nanoluc-Strep-PolyA (F182), BAP-Nanoluc+A3C5 (F183),Sumo-PAS-Lysin (F184) and IRES-BirA + polyA (F185)
Procedure:
- Minipreps were performed according to the manufacturer's protocol.
- ~200- 500ng of ach Plasmid (accept P241) were digested with EcoI and PstI for 45 min. and a gel-elctrophoresis (1% Agarose) of the digestion-batches was performed.
Results:
* P237: expected band for CMV-IgKappa+Nanoluc-Strep-PolyA at ~1850 Bp , but unexpected band at ~1100. Discard sample? * P238: expected band for CMV-IgKappa+Nanoluc-Strep-PolyA at ~1850 Bp * P239 and P240: expected band at ~650 Bp * P242 and P243: bands at ~2000 Bp. Digested vector and Fragment are not distinguishable --> sequencing
Cloning constructs for the eukaryotic and prokaryotic receptor
Investigator: CG
Aim of experiment:
Cloning of EGFR-TMD-mRuby-Strep-IRES-BirA-KDEL-PolyA, BAP-Nanoluc-A3C5-EspP-2xTerm
Procedure:
Digestion of 3 µg of each plasmid in 30 µl mix:
- 6 µl P242 (IRES-BirA-KDEL-PolyA) with EcoRI, XbaI -> Gel 1, left lane, expected fragment at about 4000 bp -> F190
- 20 µl P108 (EspP-Termin.) with EcoRI, NgoMiV -> Gel 1, right lane, expected fragment at about 3000 bp -> F191
Ligation in 20 µl mix (s. fragment names above):
- F192: 4 µl F190 + 6 µl F179
Transformation according to SOP.
Results:
Gel 1:
Transformation of P22; P67; P122; P137; P160
Investigator: MT
Aim of the experiment:
Amplification of P22; P67; P122; P137; P160
| # | Plasmid | Description |
| P22 | K157001 | RFP-Gen.(Retrafo P4) |
| P67 | pSB1C3 | EGFR signal peptide (transloc. pore) |
| P122 | pSB1C3 | Ig Kappa (digest. GSY3) |
| P137 | pSB1C3 | T7/BM40 aus GSY |
| P160 | pSB1C3 | NanoLuc + StrepTag + Poly-A |
Procedure:
- Transformation following SOP(13.02.2013)
Fermentation
Investigator: NA
Aim of the experiment: Streptavidin production
Procedure:
- exponential feed (glucose, MgSO4, Thiamin, Amp, HCDF salt, micronutrients) for ~13 h (about 3-4 L)
- Induction of streptavidin expression (anhydro tetracyclin) at OD= 111.2
- production over night
Thursday, August 11
Minipreps and analytical digestion of BBa_I0500 (AraC+ pBAD), BBa_J61046 (LoxP) and F189 (BirA)
Investigator: LK
Aim of the experiment: Isolating and verifiing the plasmids.
Procedure:
- The Preperation was performed according to the manufacturer's protocol.
- Each plasmid was digested with EcoRI and PstI and a gel-elctrophoresis (1% Agarose) of the digestion-batches was performed.
Results:
- BBa_I0500 (AraC+ pBAD) = P246 and P247: Band at ~1250; Cloning was succesful
- BBa_J61046 (LoxP) = P244 and P245: Fragment is too small (34Bp), not visible on gel.
- F189 (BirA) = P248: Band at ~ 1000Bp; Cloning was succesful
Cloning of Streptavidin-variants into pASK111
Investigator: JB, LK, CG
Aim of experiment:
Cloning of Streptactin, Traptavidin, Streptavidin into CamR pASK111
Procedure:
Digestion of about 2.5 µg of each plasmid in 30 µl mix:
- 25 µl P233 (Streptavidin) with XbaI, HindIII -> Gel 1, left lane, expected fragment at about 400 bp -> F195
- 25 µl P231 (Traptavidin) with XbaI, HindIII -> Gel 1, right lane, expected fragment at about 400 bp -> F196
- 15 µl P241 (PAS-Lysin) with XbaI, HindIII -> Gel 2, left lane, expected fragment at about 400 bp -> -
- 25 µl P229 (Streptactin) with XbaI, HindIII -> Gel 2, left right, expected fragment at about 400 bp -> F197
- 15 µl P249 (pASK111) with XbaI, HindIII -> Gel 3, left lane, expected fragment at about 3000 bp -> F198
Ligation in 20 µl mix overnight (s. fragment names above):
- F199: 3.7 µl F195 + 6.3 µl F198
- F200: 5.4 µl F196 + 4.6 µl F198
- F201: 4.1 µl F197+ 5.9 µl F198
Transformation according to SOP.
Results:
Gel 1:
Gel 2:
Gel 3:
Testpolymerization: BSA-coating
Investigator: EU, JH
Aim of experiment:
Coating of 6-well plate with BSA according to a modified ELISA-protocol.
Procedure:
Bicarbonate/carbonate coating buffer (100mM): 30,3 mg Na2CO3 60 mg NaHCO3 10 ml distilled water
"Blocking" Buffer: 1% BSA in PBS
Each well was covered with 0,5 ml BSA buffer and 0,5 ml bicarbonate buffer. After overnight incubation at RT, solution in wells was aspirated one by one and streptavidin was added. After a short incubation, the streptavidin(&Tris/HCl) solution was aspirated and Biotin solution was added (by needle).
1. well: 200 µl Streptavidin (11,2 mg/ml) + 200 µl Biotin, 2. well: 400 µl Streptavidin and 400 µl Tris/HCl + 200 µl Biotin, 3. well: 500 µl Streptavidin and 500 µl Tris/HCl + 300 µl Biotin, 4. well: 500 µl Streptavidin + 10 µl Biotin (this time with 2,5 µl pipette)
Results:
Overall polymerization was observed again, although it seemed more like an aggregation than a polymerization. No coating was observed except in the 4. well. Biotin-injection speed may be the critical point.
Poly-D-Lysine coating will follow as next step.
Transformation of P64; P73
Investigator: MT
Aim of the experiment:
Amplification of P22; P67; P122; P137; P160
| # | Plasmid | Description |
| P64 | pSB1C3 | VEGF-p2a-PDGF |
| P73 | pSB1C3 | Poly-A |
Procedure:
- Transformation following SOP(13.02.2013)
Over night liquid culture of P22; P67; P122; P137; P160
Investigator: EF, MP
Aim of the experiment:
Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
harvesting streptavidin
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- OD= 174
- centrifuge culture
- freeze pellet
SDS PAGE Streptavidin fermentation
Investigator: NA
Aim of the experiment: Verification of successfull production
Procedure:
- according to protocol
- 8 µl Ladder; before induction; after induction
- both samples: 3 µl culture + 77 µl TB + 20 µl SDS buffer; 12 µl on SDS gel
Results:
- The fermentation was not successful, streptavidin band (unter 15 kDa marker) is just a bit thicker. The reason might be the ampR of the transformed plasmid. Due to the high cell concentration beta lactamase (secreted in periplasma) gets in the medium and hydrolyses amp. As a result cells lose plasmid because there is no selective pressure without an antibiotic in medium.
- next step: cloning pf streptavidin mutants into pASK111 (camR)
Sequencing of P238, P245, P248
Investigator: NA
Procedure:
- Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) - all samples: VF2
- barcodes in stock list
Friday, August 12
PCR of Gensythesis (GSY) 7 and 8
Investigator: MT
Aim of the experiment:
Amplification of GSY 7 and 8
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 25 µL reaction | Final concentration |
| 5X Q5 Reaction Buffer | 5 µL | 1 X |
| 10 mM dNTPs | 0.5 µL | 200 μM |
| 10 μM Forward Primer (D20) | 1.25 μl | 0.5 μM |
| 10 μM Reverse Primer (VR) | 1.25 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Q5 High-Fidelity DNA Polymerase | 0.25 μl | 0.02 U/μl |
| Nuclease-Free Water | to 25 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 5 - 10 sec |
| 61 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 16 °C |
Note: Use of the NEB T m Calculator is highly recommended.
Results:
After running on 1 %agarose gel only the 2kb bands belonging to GSY8 could be distinguished. These were extracted from the gel.
-> Repeat PCR for GSY7
Gelextraction of GSY8
Investigator: EF
Aim of the experiment: Gelextraction of 2kb band of PCR belonging to GSY8 Procedure:
Gelextraction was performed by manufacturers protocol (SLG).
Concentration:
7,7 ng/ul
Plasmid preparation of liquid culture of P22; P67; P122; P137; P160
Investigator: MP
Aim of the experiment:
Purification of P22; P67; P122; P137; P160
| # | Plasmid | Description |
| P22 | K157001 | RFP-Gen.(Retrafo P4) |
| P67 | pSB1C3 | EGFR signal peptide (transloc. pore) |
| P122 | pSB1C3 | Ig Kappa (digest. GSY3) |
| P137 | pSB1C3 | T7/BM40 aus GSY |
| P160 | pSB1C3 | NanoLuc + StrepTag + Poly-A |
Procedure:
- According to Mini-prep kit, SLG
Results:
- P22 ->P1001 103.6 ng/µL
- P67 ->P1004 92.9 µL
- P122 -> P1000 80.8 ng/µL
- P137 -> P1003 87.9 µL
- P160 -> P1002 100 ng/µL
Dephosphorylation of F113[HincII digested]
Investigator: MP, EF
Aim of the experiment:
Dephosphorylation of F113[HincII digested] (RFP-Reportercassette )
Procedure: According to NEB online Protocol for Dephosphorylation of 5 ́-ends of DNA using Antarctic Phosphatase (M0289):
- 12 µL F113 (85.1 ng/µL)
- 2 µL Antarctic Phosphatase Rxn Buffer (10X)
- 1 µL Antarctic Phosphatase (5000 u/mL)
- 5 µL ddH2O
Midiprep P238 (CMV-IgKappa-Nanoluc-Strep-polyA)
Investigator: NA
Procedure:
- MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen)
Results:
P253 - concentration: 2134 ng/µl
=== Digestion, Gel extraction and ligation of F1000 (P122), F1001 (P67), F1002 (P160) and F1003 (P137) ===
Investigator: EU
Aim of the experiment:
Preparation of plasmids
Procedure:
- 33 µL DNA
- 1 µL enzyme 1
- 1 µL enzyme 2
- 5 µL cut smart buffer
- 10 µL ddH20
Enzymes used:
- F1000: AgeI + PstI
- F1001: AgeI + PstI
- F1002: NgoMiT + PstI
- F1003: AgeI + PstI
Gelextraction:
- Using gelextraction kit, Quiagen
Ligation
- 2µl F1000 + 6µl F1002
- 2µl F1001 + 6µl F1002
- 2µl F1003 + 6µl F1002
+ 1µl T4 Ligase and 1 l T4 ligase buffer for each ligation
Results:'
Left to right F1002(2x), F1001(2x), F1000(2x) and F1003(1x)
- F1000 30 ng/µL
- F1001 23,8 ng/µL
- F1002 9,4 ng/µL
- F1003 23,1 ng/µL
Saturday, August 13
Miniprep of pASK111 (empty) and diegestion
Investigator: LK
Aim of the experiment: Amplification of pASK111 and digestion for further ligation with SUMO-PAS-Lysin.
Procedure:
- The Miniprep was performed according to the manufacturer's protocol.
- The whole DNA was diegested with XbaI and HindIII. Over night at 37°C.
- Gelelectrophoresis was performed and showed a significant band at ~2700.
Comment
- Gel-ectraction has yet to be done .
- equally digested insert from P236 was visible on gel. --> no ligation possible.
Transformation of F1006; F1007; F1008
Investigator: MT
Aim of the experiment:
Amplification of F1006; F1007; F1008
| # | Ligation | Plasmid | Description |
| F1006 | F1000 + F1002 | pSB1C3 | Ig Kappa + NanoLuc + StrepTag + Poly-A |
| F1007 | F1003 + F1002 | pSB1C3 | T7/BM40 + NanoLuc + StrepTag + Poly-A |
| F1008 | F1001 + F1002 | pSB1C3 | EGFR + NanoLuc + StrepTag + Poly-A |
Procedure:
- Transformation following SOP(13.02.2013)
Digestion and gelextraction of P64; P73
Investigator: JB, MT
Aim of the experiment:
Preparation for ligation
Procedure:
- 10 µL DNA
- 1 µL enzyme 1
- 1 µL enzyme 2
- 2 µL buffer
- 6 µL ddH20
Enzymes used:
- P64: EcoRI + SpeI
- P73: EcoRI + XbaI
Gelextraction:
- Using gelextraction kit, Quiagen
Results:'
Left: P64 Right: P73
- P64 -> F1011 14,2 ng/µL
- P73 -> F1012 17,6 ng/µL
Sunday, August 14
Miniprep of P199-P201 and analytical digestion
Investigator: LK
Aim of the experiment: Isolation of "Streptavidin-variants" in pASK111 for preinoculation tomorrow.
Procedure:
- The Miniprep was performed according to the manufacturer's protocol.
- ~200ng where digested with XbaI and HindIII.
Results:
- All samples showed significant bands at ~400 Bp.
Transformation of P199-201 into E.coli BL-21
Investigator: LK
Aim of the experiment: Introducing Strep-variants in pASK111 into BL-21 for further fermentation.
Procedure:
- The cells were transformed according to the SOP with 1µl of DNA and after 1h of incubation in LB without antibiotics plated out on Cam-plates.
Miniprep of P113-P213 and analytical digestion
Investigator: LK Aim of the experiment: Isolation of "Streptavidin-variants" in pASK111 for preinoculation tomorrow.
Procedure:
- The Miniprep was performed according to the manufacturer's protocol.
- ~200 ng were digested with EcoRI and PstI.
Results:
PCR of Gensynthesis (GSY) 7, 8
Investigator: MT
Aim of the experiment:
Amplification of GSY7
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491)
- Different amounts of template DNA were used
| Component | 50 µL reaction | Final concentration |
| 5X Q5 Reaction Buffer | 10 µL | 1 X |
| 10 mM dNTPs | 1 µL | 200 μM |
| 10 μM Forward Primer (D20) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (VR) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 1 - 10 ng |
| Q5 High-Fidelity DNA Polymerase | 0.5 μl | 0.02 U/μl |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 40 cycles | 98 °C | 5 - 10 sec |
| 58 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Results:
- PCR GSY8 successful.
-> Do it again for GSY 7!
Over night liquid culture of F1006; F1007; F1008
Investigator: MT
Aim of the experiment:
Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Gelextaction of F1004
Investigator: MT
Aim of the experiment:
Procedure:
- Using gelextraction kit, Quiagen
Results:'
- long F1004 -> F1013 6,4 ng/µL
- short F1004 -> F1014 1,1 ng/µL
Ligation F1012 + F1011
Investigator: MT
Aim of the experiment:
Ligation F1012 + F1011
| # | Plasmind | Description |
| F1011 | VEGF-p2a-PDGF | |
| F1012 | pSB1C3 | Poly-A |
Procedure:
- 3 µL F1012
- 6 µL F1011
- 2 µL buffer
- 1 µL T4 Ligase
- 8 µL ddH2O
Results:
- F1015
Transformation of F1015
Investigator: MT
Aim of the experiment:
Amplification of F1015
| # | Ligation | Plasmid | Description |
| F1015 | F1011 + F1012 | pSB1C3 | VEGF-p2a-PDGF + poly-A |
Procedure:
- Transformation following SOP(13.02.2013)
Results:
- No colonies. Do it again.
Week 14, Aug 15th - Aug 21st
Monday, Aug 15th
Analytical digestion of P251 and P252
Investigator: Clemens
Aim of the experiment: Verify cloning
Procedure:
- 1µL Plasmid was digested with 1µL of EcoRI and PstI
- Digestions were incubated for 1h
- Samples were seperated by gelelectrophoresis on a 1% agarose gel
Results:
- Cloning seems to have worked
Plasmidpreparation of liquid culture of F1006; F1007; F1008
Investigator: MP
Aim of the experiment:
Purification of F1006; F1007; F1008
| # | Plasmid | Description |
| F1006 | pSB1C3 | Ig Kappa + NanoLuc + StrepTag + Poly-A |
| F1007 | pSB1C3 | T7/BM40 + NanoLuc + StrepTag + Poly-A |
| F1008 | pSB1C3 | EGFR + NanoLuc + StrepTag + Poly-A |
Procedure:
- According to Mini-prep. kit, Quiagen
Results:
- F1006 -> F1005 83.2 ng/µL
- F1007 -> F1006 91.8 ng/µL
- F1008 -> F1007 367.1 ng/µL
Cloning of Nanoluc-Strep in front of double terminator and PAS-Lysine into pASK111
Investigator: LK, CG
Aim of experiment:
Cloning of Nanoluc-Strep in front of double terminator and PAS-Lysine into pASK111
Procedure:
Plasmid preperation according to SOP of P264, P265
Digestion of about 3 µg of each plasmid in 30 µl mix:
- 6 µl P29 (double teminator) with EcoRI, XbaI -> Gel 1, left lane, expected fragment at about 3000 bp -> F207
- 15 µl P98 (Nanoluc-Strep) with EcoRI, SpeI -> Gel 1, right lane, expected fragment at about 500 bp -> F208
- 15 µl P264 (PAS-Lysin) with XbaI, HindIII -> Gel 2, left lane, expected fragment at about 1000 bp -> F210
Ligation in 20 µl mix for 30 min. at room temperature (s. fragment names above):
- F209: 3.7 µl F207 + 6.3 µl F208
- F211: 1.1 µl F210 + 8.9 µl F206
Transformation according to SOP.
Results:
Gel 1:
Gel 2:
Tuesday, Aug 16th
Over night liquid culture of P64; P73
Investigator: MP
Aim of the experiment:
Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Gelextraction of PCR GSY8(Saturday, August 13)
Investigator: EF
Aim of the experiment:
Purification of PCR GSY8(Sunday, August 14)
Procedure:
- Using gelextraction kit, SLG
Results:
- F1016 3,4 ng/µL
Seeding of HEK293T cells for transfection
Investigator: LK, CG
Aim of the experiment:
3x4 petri dishes with HEK293T cells for performing the Nanoluc assay in order to validate the signale peptides
Procedure:
- Washing each T75 flask with 10 ml PBS
- Centrifugation for 10 min. at 1.8 rcf
- Resuspending the cell pellet in 10 ml medium
- Counting the cells (33x10^6 cells in 10 ml)
- Plating 0.5x10^6 cells onto petri dishes in 10 ml medium an 1x10^6 onto a T75 flask
- Incubation for 2 days
Results:
- ready to transfect cells in petri dishes
Gelextraction of PCR GSY8(Saturday, August 13)
Investigator: MP
Aim of the experiment:
Purification of PCR GSY8(Sunday, August 14)
Procedure:
- Using gelextraction kit, SLG
Results:
- F1016 3,4 ng/µL
Transformation of F1015
Investigator: MT
Aim of the experiment:
Amplification of F1015
| # | Ligation | Plasmid | Description |
| F1015 | F1011 + F1012 | pSB1C3 | VEGF-p2a-PDGF + poly-A |
Procedure:
- Transformation following SOP(13.02.2013)
PCR of Gensynthesis (GSY) 7
Investigator: EF
Aim of the experiment:
Amplification of GSY7
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491)
- Reactions with and without GC Enhancer were performed
| Component | 50 µL reaction x 6 | Final concentration |
| 5X Q5 Reaction Buffer | 10 µL | 1 X |
| 10 mM dNTPs | 1 µL | 200 μM |
| 10 μM Forward Primer (D20) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (VR) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 1 ng |
| Q5 High-Fidelity DNA Polymerase | 0.5 μl | 0.02 U/μl |
| 5X Q5 High GC Enhancer (optional) | 5 µL | 1X |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 40 cycles | 98 °C | 5 - 10 sec |
| 58 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 16 °C |
Results:
- negativ. Two 1% agarose gels were performed to prove the PCR product. Both were negativ.
Precipitation of eGFP
Investigator: LK
Aim of experiment: Determination of (NH4)2SO4 -concentration needed to precipitate eGFP.
Procedure:
- 50mL of eGFP solution (from fermentation) where gradually precipitated with rising concentrations (NH4)2SO4 (step size: 400mM)
- All steps where performed at 4°C.
- Pellets where solved in 10 mL 1x PBS.
Results:
Extinction:
- Maximum Absorption at 488nm for eGFP. Absorption coefficient(eGFP,488): 53000 mol/Lcm.
| mM (NH4)2SO4 | Dilution | Extinction | Concentration eGFP [µM] |
| 400 | 1:10 | 0,197 | 37,0 |
| 800 | 1:10 | 0,084 | 15,8 |
| 1200 | 1:10 | 0,457 | 86,2 |
| 1600 | 1:100 | 0,16 | 301,8 |
| 2000 | 1:100 | 0,375 | 707,5 |
| 2400 | 1:10 | 0,505 | 95,3 |
| 2800 | 1:10 | 0,08 | 15,0 |
Inocculation of P258_E.coli_Bl-21 in M9 medium for fermentation
Investigator: LK
Procedure:
- M9 medium with Cam (1:1000) was inocculated with preculture (~50 mL) of P258 (pASK111+Streptavidin)
- the culture was incubated in the shaker over night at 37°C
Phosphorylation of F1005; F1016
Investigator: MP
Aim of the experiment:
Phosphorylation of F1005; F1016
Procedure:
- 39 µL DNA
- 5 µL 10x T4 PNK Bufer
- 10 mM ATP
- T4 PNK
Phosphorylation of O31; O32
Investigator: MP
Aim of the experiment:
Phosphorylation of O31; O32
Procedure:
- 0.5 µL Oligonucleotides (10µM)
- 2 µL 10x T4 PNK Bufer
- 2 µL 10 mM ATP
- 1 µL T4 PNK
- 14.5 µL H2O
- 95 °C; 3 min
Hybridization of phosphorylated O31; O32
Investigator: MP
Aim of the experiment:
Hybridization of phosphorylated oligonucleotides: O31; O32
Procedure:
- 2 µL O31
- 2 µL O32
- 200 µL ddH20
- 37 °C 20 min
- 75 °C 10 min
Wednesday, August 17
Gelelectrophoresis of GSY7
Investigator: EF
Aim of experiment: gelelectrophoresis of the remaining product of GSY7 PCR (from 16.8.16)
Procedure: Agarose gelelectrophoresis was performed using an increased amount of SYBR Green
Results There was no PCR product identified.
Plasmidpreparation of liquid culture of P64; P73
Investigator: MP
Aim of the experiment:
Purification of P64; P73
| # | Plasmid | Description |
| P64 | pSB1C3 | VEGF-p2a-PDGF |
| P73 | pSB1C3 | Poly-A |
Procedure:
- According to Mini-prep. kit, SLG
Results:'
- P64 134.4 ng/µL
- P73 149.4 ng/µL
Over night liquid culture of F1015
Investigator: MP
Aim of the experiment:
Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
PCR of scAvidin
Investigator: Julian
Aim: Extraction of single chain Avidin from vector for further cloning
Procedure:
- reaction was performed after SOP (PCR Using Q5® High-Fidelity DNA Polymerase, NEB)
- 2 ng Plasmid were used for amplification (1 µL)
- Primers: O-27 and O-28
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial denaturation | 98 | 120 |
| 35 cycles | 98 | 10 |
| 64 | 30 | |
| 72 | 300 | |
| final extension | 72 | 300 |
| Hold | 4 | ꝏ |
Quickchange of P206 and P239 to correct BAP
Investigator: Julian
Aim: Quickchange P206 (BAP-NanoLuc) and P239 (BAP-NanoLuc-A3C5) to correct BAP
Procedure:
- reaction was performed after SOP (PCR usingPfuUltra II Fusion HS DNA Polymerase, Agilent)
- 10 ng Plasmid were used for amplification (2.5 µL)
- Primers: O-33 and O-34
| Step | Temperature [°C] | Time [s] |
| initial denaturation | 98 | 120 |
| 35 cycles | 98 | 10 |
| 55 | 30 | |
| 68 | 300 | |
| final extension | 68 | 360 |
| Hold | 4 | ꝏ |
Inocculation of fermenter
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- centrifuge preculture (P258 in M9 medium)
- resuspend pellet in HCDF medium of fermenter and inocculate
- pO2= 60 %; Temp= 37°C, pH~7
- growth over night
Thursday, Aug 18th
PCR of gene synthesises 8 with Q5 polymerase
Investigator: EF
Aim of the experiment: Amplification of GSY7 with optimized Tm following negativ PCR reactions in order to amplify GSY7. Procedure: Approach:
- 2,5 µl forward Primer VF2
- 2,5µl reverse Primer VR
- 1µl dNTPs
- 1µl DNA (1ng)
- 10µl 5x Q5-Puffer
- 0,5µl Q5-Polymerase
- Fill up with water up to 50 µl
Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/)
- 98°C_2min initiation step
- 98°C_10sec
- 62°C_30sec > step 2-4: 35 repeats
- 72°C_1min
- 72°C_5min final step
- 4°_hold
PCR of gene synthesises 8 with Pfu Ultra II polymerase
Investigator: EF
Aim of the experiment: Amplification of GSY7 with Pfu Ultra II polymerase following negativ PCR reactions in order to amplify GSY7. Procedure: Approach:
- 2,5 µl forward Primer VF2
- 2,5µl reverse Primer VR
- 1µl dNTPs
- 1µl DNA (1ng)
- 10µl Pfu Ultra II-Puffer
- 0,5µl Q5-Polymerase
- Fill up with water up to 50 µl
Setup: (calculated with NEB calc. http://tmcalculator.neb.com/#!/)
- 98°C_2min initiation step
- 98°C_10sec
- 56°C_30sec > step 2-4: 35 repeats
- 72°C_2min
- 72°C_5min final step
- 4°_hold
Fermentation
Investigator: NA
Aim of the experiment: Streptavidin production
Procedure:
- exponential feed (Cam) for ~13 h (about 3-4 L); start at OD= 11.66
- Induction of streptavidin expression (anhydro tetracyclin) at OD= 119
- change of settings: pO2= 40 %; Temp= 30°C, pH~7
- production over night
Streptavidin production in shaking flask
Investigator: NA
Aim of the experiment: Streptavidin production
Procedure:
- 2 L LB medium (Cam 1:1000) was inocculated with ~10 ml M9 preculture (P258 in BL21)
- induction with anhydro tetracycline (200µl, 2 mg/ml) at OD= 3,32
- culture was incubated at 37 °C and 140 rpm for 4 hours
- after expression, cultures were transferred into centrifuge tubes and spun down in the centrifuge (4 °C, 5000 rpm, 20 min, F4X1L rotor)
- the supernatant was discarded, and the pellet was frozen away
Transformation of quickchanged P239 and 206 (BAP_Nanoluc_A3C5 and BAP_Nanoluc)
Investigator: LK
Procedure:
- After DpnI-digestion 10µl of each plasmid was transformed into E.coli XL-1-blue.
- Cells where plated out an Cam-plates after 1h of incubation without antibiotics.
MiniPrep of F209 to P266 and P267, analytic digestion
Investigator: JL
Aim of the experiment: Verification of successful cloning.
Procedure: MiniPrep was performed according to manufacturer's protocol (QIAprep MiniPrep, Qiagen)
- Concentrations:
| Plasmid | c [ng/µl] |
| P266 | 198 |
| P267 | 200,8 |
- Analytic digestion: 0.5 µl PstI, 0,5 µl EcoRI, 1 µl CutSmart, 6 µl ddH2O for P266 and P267 for 2 h at 37°C.
- 1% agarose gel electrophoresis.
Results:
- Lane 1: 10 µl 1 kb Ladder.
- Lane 2: Digest of F209 to P266.
- Lane 3: Digest of F209 to P267.
Phosphorylation of O31; O32
Investigator: MP
Aim of the experiment:
Phosphorylation of O31; O32
Procedure:
- 5 µL Oligonucleotides (µM)
- 2 µL 10x T4 PNK Bufer
- 2 µL 10 mM ATP
- 1 µL T4 PNK
- 10 µL H2O
- 95 °C 3min.
Hybridization of phosphorylated O31; O32
Investigator: MP
Aim of the experiment:
Hybridization of phosphorylated oligonucleotides: O31; O32
Procedure:
- 2 µL O31
- 2 µL O32
- 200 µL ddH20
Plasmidpreparation of liquid culture of F1015
Investigator: MP
Aim of the experiment:
Purification of F1015
| # | Plasmid | Description |
| F1015 | pSB1C3 | VEGF-p2a-PDGF + Poly-A |
Procedure:
- According to Mini-prep. kit, SLG
Results:'
- F1015 -> P1008 256.6 ng/µL
Friday, Aug 19th
Gelelectrophoresis of GSY7
Investigator: EF
Aim of experiment: gel electrophoresis of repeated GSY7 PCR (from 18.8.16)
Procedure: Agarose gelelectrophoresis was performed at 90V
Results There was no PCR product identified.
harvesting streptavidin
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- OD= 181
- centrifuge culture for 1 h at 4800 rpm
- freeze pellet (2033 g)
SDS PAGE Streptavidin fermentation
Investigator: NA
Aim of the experiment: Verification of successfull production
Procedure:
- according to protocol
- 8 µl Ladder; before induction; after induction; sample of shaking flask production after induction
- all samples: 0.5/OD µl culture filled up with EB to 80µl + 20 µl SDS buffer; 10 µl on SDS gel
Results:
Clear bands of acetyl transferase (chloramphenicol resistance) in fermenter samples. Therefore cells kept plasmid. The streptavidin bands before and after induction do not differ. The fermentation was not succesful. The shaking flask sample however shows a brighter band for streptavidin.
Miniprep of F211 (pASK111(PAS Lys)), I739001 (Biobrick) and F206 (pASK111 vectorfragment)
Investigator: Clemens
Aim of the experiment: Miniprep of plasmids Procedure:
- Miniprep was performed by manufacturers protocol (Qiaprep, Qiagen)
- Plasmids were named:
- F211 a: P268
- F211 b: P269
- I739001 a: P270
- I739001 b: P271
- F206 a: P272
- F206 b: P273
Cloning of P260 in P261 and F215 in P74
Investigator: Clemens
Aim of the experiment: Digestion, ligation and transformation Procedure:
Transfection of HEK cells with P253, P225 and P226
Investigator: LK
Aim of the experiment: Transfecting HEK cells with vectors encoding 3 diffrent types of signal peptides for secretion.
Procedure:
- HEK293 where cultured and seeded out on 12 cellculture dishes. (~5x105 cells per dish)
- Transfection was performed on day 3 after seeding (60-70% confluency)
- 10µg DNA of each signalpeptide mixed with 30µg PEI (polyenthylenimine) in ddH2O where incubated for 15 min and added to the cells, which where previously washed with PBS and covered (4mL) with optiMEM media. (Pen/Strep;No serum!)
- After 4h of incubation at 37°C, 5% CO2 the media was changed to RPMI (Pen/Strep; 5% FCS) again.
Picking of E. coli XL1 blue in quickchanged pSB1C3 in P239 and P206, Retrafo of P261 and P213
Investigator: JL
Procedure:
- Quickchanged pSB1C3 in P239: Colonies were picked from Cam-plates.
- Retrafo of P261 and P213: Colonies were picked from Cam-plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL of the proper antibiotic.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Repeat of PCR of scAvidin
Investigator: Julian
Aim: Extraction of single chain Avidin from vector for further cloning
Procedure:
- reaction was performed after SOP (PCR Using Q5® High-Fidelity 2X Master Mix, NEB)
- 2 ng Plasmid were used for amplification
- Primers: O-27 and O-28
- annealing temperature was lowered to 60 °C
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial denaturation | 98 | 120 |
| 35 cycles | 98 | 10 |
| 60 | 30 | |
| 72 | 150 | |
| final extension | 72 | 300 |
| Hold | 4 | ꝏ |
Repeat of PCR of Gene Synthesis 9
Investigator: Julian
Aim: Amplification of gensynthesis 9 for further cloning
Procedure:
- reaction was performed after SOP (PCR Using Q5® High-Fidelity 2X Master Mix, NEB)
- 2 ng Plasmid were used for amplification
- Primers: VF2 and VR
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial denaturation | 98 | 120 |
| 35 cycles | 98 | 10 |
| 61 | 30 | |
| 72 | 120 | |
| final extension | 72 | 300 |
| Hold | 4 | ꝏ |
Preparative digestion of P8 (Streptavidin wt)
Investigator: Julian
Aim of experiment: preparation for cloning of SA1 into pASK111
Procedure:
- preparative digestion batch
| volume | reagent |
| 40 µl | Plasmid DNA P8 |
| 5 µl | Cutsmart (10x) |
| 1.5 µl | HindIII |
| 1.5 µl | XbaI |
| 2 µl | ddH2O |
| =50 µl | TOTAL |
- duration: over night
Saturday, Aug 20th
Miniprep and analytical digestion of P274-P277
Investigator: LK
Aim of the experiment: Isolating and verifiing A3C5+EspP_ddT ligation, "quickchanged" BAP-constructs and amplification of A3C5 in pSB1C3.
Procedure:
- Minipreps where performed according to manufacturer's protocol.
- Analytical digestion with EcoRI and PstI of ~ 200-250ng of each plasmid was performed for 30 min at 37°C.
Results:
Sunday, Aug 21st
Cloning of GSY9 and scAvidin
Investigaor: LK, JL
Procedure:
- The PCR of scAvdin was digested over night with SpeI and NgoMIV at 37°C.
- The PCR of GSY9 was divided: one half was digested with XbaI and AgeI (-> F224) and the other half was digested with XbaI and HindIII (->F225).
- Additionally P74 (pSB1C3+RFP-Reporter) was digested with XbaI and AgeI (-> F227) to provide a plasmid for the ligation with F224.
- The digestions where plotted on a gel, and bands at 1700Bp for scAvidin, 222Bp for TetO-OmpA, 490Bp for eMA and 2100Bp for pSB1C3 where cut out and purified with the Promega Wizard SV Gel Kit according to the manufactuerers protocol.
- F224 (TetO-OmpA) was then ligated with F227 (pSB1C3), F225 (enhanced monomeric Avidin) was ligated with F206 (pASK111) and F226 (scAvidin) was ligated with F218 (pSB1C3). Ligations where incubated over night at 16°C.
- Transformation was performed according to the SOP on Monday morning.
Week 15, Aug 22nd - Aug 28th
Monday, Aug 22nd
Sequencing of P268, P276, P277 and P279
Investigator: Julian
Aim of the experiment: Sequencing of P268 (PAS lysine), P276 (QC BAP-NanoLuc-A3C5), P277 (QC BAP-NanoLuc) and P279 (A3C5-EspP-DoubleTerminator)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
- P268 (D20) FR12904429
- P276 (VF2) FR12904428
- P277 (VF2) FR12904427
- P279 (VF2) FR12904426
Analytical digestion of P278 (pSB1C3(A3C5-EspP-doubleterminator) & P279
Investigator: Clemens
Aim of the experiment: cofirm success of cloning
Procedure:
- Batch for analytical digestion for P278/P279 with PstI-HF and EcoRI-HF
| volume | reagent |
| 2 µl/ 1 µl | Plasmid DNA P278/P279 |
| 1 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 5 µl/ 6 µl | ddH2O |
| =10 µl | TOTAL |
Results:
- P279 showed the expected signal at 1100 bp
Ligations of F1013 + gsy7; +gsy8
Investigator: EF
Aim of the experiment: cloning for further ligation steps
Procedure:
Tuesday, Aug 23rd
Inoculation of Precultures of F228, F229 and F230
Investigator: Clemens
Aim of the experiment: Prepare cells for Miniprep
Procedure:
- Two colonies were picked from each plate
- LB CAM was inoculated with F228 and F230
- LB AMP was inoculated with F229
- Precultures were incubated for 7 hours at 37°C
Results:
- Each preculture showed cell growth
Miniprep of F228, F229 and F30
Investigator: Clemens
Aim of the experiment: Obtain plasmids
Procedure:
- Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN)
- Plasmids were renamed:
- F228 1: P281
- F228 2: P282
- F229 1: P283
- F229 2: P284
- F230 1: P285
- F230 2: P286
Results:
- Concetrations in "Inventarliste"
Digestion of IRES-construct (F1019) by PmeI+NotI
Investigator: EU
Aim of the experiment: Prepare construct for ligation later on
Procedure:
5 µl DNA 38µl ddH2O 1µl of each enzyme 5µl cut smart buffer
incubation at 37°C for 1 h Gel electrophoresis and gel extraction
Results:
Fragment was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder.
Wednesday, Aug 24th
analytical digestion of P281, P282, P283, P284, P285 and P286
Investigator: Clemens
Aim of the experiment: Verify success of cloning
Procedure:
- Batch for analytical digestion of P281/P282 with PstI-HF and EcoRI-HF
| volume | reagent |
| 1 µl/ 1 µl | Plasmid DNA P281/P282 |
| 1 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 6 µl/ 6 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P283/P284 with XbaI and HindIII-HF
| volume | reagent |
| 1 µl/ 1 µl | Plasmid DNA P283/P284 |
| 1 µl | Cutsmart buffer (10x) |
| 1 µl | XbaI |
| 1 µl | HindIII-HF |
| 6 µl/ 6 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P285/P286 with PstI-HF and EcoRI-HF
| volume | reagent |
| 1 µl/ 1 µl | Plasmid DNA P285/P286 |
| 1 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 6 µl/ 6 µl | ddH2O |
| =10 µl | TOTAL |
- Digestions were incubated at 37° for 1h
Results:
PCR of Genesynthesis 9 with VF2 and VR
Investigator: Clemens
Aim of the experiment: Amplification of GSY9
Procedure:
- reaction was performed after SOP (Q5 Hotstart, NEB)
- 2 ng Plasmid were used for amplification
- Primers: VF2 and VR
- Over all volume= 50 µl
- PCR program (standard):
| Step | Temperature [°C] | Time [s] |
| initial denaturation | 98 | 120 |
| 35 cycles | 98 | 10 |
| 66 | 30 | |
| 72 | 30 | |
| final extension | 72 | 300 |
| Hold | 4 | ꝏ |
- PCR purification was performed according to manufacturers protocol (Wizard SV Gel and PCR clean-up kit, Promega)
Results:
PCR of linear pSB1C3
Investigator: MT
Aim of the experiment:
Amplification of linear pSB1C3 and introduction of restriction site: blunt at 5' and NotI at 3'
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (O-038) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (O-039) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 1 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 10 sec |
| 68 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Preparative digestion of P74, P165, P196, P273, P274, P275, P285 and PCR of GSY9
Investigator: Clemens
Aim of the experiment:
Procedure:
- Batch for preparative digestion of P74 with XbaI and AgeI-HF
| volume | reagent |
| 2 µl | Plasmid DNA P74 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | XbaI |
| 1 µl | AgeI-HF |
| 23 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P74 with NgOMIV and AgeI-HF
| volume | reagent |
| 2 µl | Plasmid DNA P74 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | AgeI-HF |
| 23 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P165 with PstI-HF and AgeI-HF
| volume | reagent |
| 12 µl | Plasmid DNA P165 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | AgeI-HF |
| 13 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P196 with NgOMIV and PstI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P196 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 10 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P273 with XbaI and HindIII-HF
| volume | reagent |
| 20 µl | Plasmid DNA P273 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | XbaI |
| 1 µl | HindIII-HF |
| 5 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P274 with AgeI-HF and PstI-HF
| volume | reagent |
| 10 µl | Plasmid DNA P274 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | AgeI-HF |
| 15 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P275 with NgOMIV and PstI-HF
| volume | reagent |
| 8 µl | Plasmid DNA P275 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 17 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P285 with NgOMIV and PstI-HF
| volume | reagent |
| 12 µl | Plasmid DNA P285 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 13 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of PCR of GS9 with NgOMIV, AgeI-HF and XbaI
| volume | reagent |
| 25 µl | Plasmid DNA PCR GSY9 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 0 µl | ddH2O |
| =31 µl | TOTAL |
- Batch for preparative digestion of PCR of GS9 with HindIII-HF and XbaI
| volume | reagent |
| 25 µl | Plasmid DNA PCR GSY9 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | XbaI |
| 1 µl | HindIII-HF |
| 0 µl | ddH2O |
| =30 µl | TOTAL |
- Digestions were incubated at 37° over night
- Samples were applied on a 1% agarose gel
- Samples were seperated for 45 min at 80V
Results:
- All Digests showed the expected lengths
- Fragments were cut from the gel and purified according to manufacturers protocol (Wizard SV Gel and PCR clean-up system, Promega)
Transformation of P296 (pSB1C3 mGMK-Tk30, BBa_K782063) in xl1-blue
Investigator: MT
Aim of the experiment: Transformation for Poly-A-tail adding
Procedure: transformation in competent xl1-blue according to protocol + rescue
Result: plates (LB Cam) in incubator for further processing (37 °C) showed after 24 h colonies
(Re)ligations of F1013 (pSB1C3) and F1016 (phosphoryliert) + F1013 and F1005 in pSB1C3
Investigator: EF
Aim of the experiment: Ligations of PCR Produkt from GSY8 in pSB1C3 vector for later transformation and cloning
Procedure:
- Ligation mixtures were prepared as following; DNA amounts were calculated using the ligation calculator http://www.insilico.uni-duesseldorf.de/Lig_Input.html :
- F1013+F1016: 1.6 μL F1013, 5 μL F1016, 2μL T4-DNA-Ligase-Buffer, 1 μL T4-DNA-Ligase, 10.4 μL ddH2O
- F1013+F1005: 1.6 μL F1013, 2.2 μL F1005, 2μL T4-DNA-Ligase-Buffer, 1 μL T4-DNA-Ligase, 13.2 ddH2O
- Ligation was incubated for 1.5 hours at 16 °C
Thursday, Aug 25th
Phosphorylation and hybridization of O35, O36, O37
Investigator: EU
Aim of the experiment:
Phosphorylation and hybridization of O35, O36, O37
Procedure: Phosphorylation
- 0,5 µL Oligonucleotides
- 2 µL 10x T4 PNK Bufer
- 2 µL T4 Ligase Buffer
- 1 µL T4 PNK
- 14,5 µL H2O
incubate at 37°C for 20 min and inactivate T4PNK (for 10 min at 75°C)
Hybridization
- 2 µL O31
- 2 µL O32
- in 200 µL ddH20
incubate at 95°C for 3 min, let cool down slowly
Results:
Inoculation of P296(Kill-Switch)
Investigator: EU
Aim of the experiment: production of P296
Procedure:
2x5ml LB-CAM media, each inoculated by one colony (not by rescue colonies)
Results:
Digestion of P74 + HincII
Investigator: MT
Aim of the experiment:
- Digestion of P74 + HincII
Procedure:
- 3 µL DNA (1431 ng/µL)
- 1 µL HincII
- 14 µL ddH2O
- 2 µL CutSmart buffer
- incubation for 2h at 37 °C
Results:'
- P74 -> P1021 111,2 ng/µL
Ligation of F233+F238, F236+F234, F233+F237, F232+F240, F231+F239, F235+F241 and F248+F249
Investigator:
Aim of the experiment:
- Ligation and transformation of XL1 blue
Procedure:
- Batch for Ligation of F233 and F238
| volume | reagent |
| 1,7 µl | Vector F233 |
| 8,3 µl | Insert F238 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F236 and F234
| volume | reagent |
| 1,1 µl | Vector F236 |
| 8,9 µl | Insert F234 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F233 and F237
| volume | reagent |
| 3,1 µl | Vector F233 |
| 6,7 µl | Insert F237 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F232 and F240
| volume | reagent |
| 4,8 µl | Vector F232 |
| 5,2 µl | Insert F240 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F231 and F239
| volume | reagent |
| 5,6 µl | Vector F231 |
| 4,4 µl | Insert F239 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F235 and F241
| volume | reagent |
| 6,5 µl | Vector F235 |
| 3,5 µl | Insert F241 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F228 and F249
| volume | reagent |
| 2,7 µl | Vector F248 |
| 7,3 µl | Insert F249 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Ligations were performed according to SOP
- XL1blue were transformed with the resulting fragments
Results:'
Cellgrowth yes/no?
Digestion of P1007 + EcoRI + SpeI
Investigator: EF
Aim of the experiment:
- Digestion of P1007
Procedure:
- 10 µL DNA (3.6 ug)
- 0.25 µL EcoRI
- 0.25 µL SpeI
- 34.5 µL ddH2O
- 5 µL CutSmart Buffer NEB
- incubation for 1h at 37 °C
Results:
Preparative digestion, gel electrophoresis and gel extraction of P288 (CMV-Promotor) and P289 (NanoLuc + Streptag + Poly-A)
Investigator: JL
Aim of experiment: Preparative digestion, gel electrophoresis and gel extraction of P288 (CMV-Promotor) and P289 (NanoLuc + Streptag + Poly-A)
Procedure:
- Batches for analytical digestions:
- P288: PstI and SpeI.
- P29: PstI and XbaI.
| volume | reagent |
| 6 µl | Plasmid DNA |
| 19 µl | ddH2O |
| 3 µl | CutSmart buffer (10x) |
| 1 µl | Each enzyme |
| 30 µl | TOTAL |
- Preparative gel electrophoresis was performed at 90 V for 60 min.
- Preperative gel according to SOP.
- The Purification was done according to the manufacturer's protocol.
Analytical digestion and gelelectrophoresis of P293, P294, P295 (eMA pASK111)
Investigator: JL
Aim of the experiment: Analytical digestion and gelelectrophoresis of P293, P294, P295 (eMA pASK111).
Procedure:
- Batch for analytical digestion for P293, P294, P295.
| volume | reagent |
| 2/1/4 µl | Plasmid DNA P293/P294/P295 |
| 2 µl | NEBuffer 4 (10x) |
| 0.5 µl | XbaI |
| 0.5 µl | HindIII (20 U/µl) |
| 6/8/4 µl | ddH2O |
| =12 µl | TOTAL |
- Incubation for 120 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 30 min.
Results:
| 1 kbp ladder DNA ladder | P293 | P294 | P294 |
Friday, Aug 26
Digestion of P74 + HincII
Investigator: MT
Aim of the experiment:
- Digestion of P74 + HincII
Procedure:
- 3 µL DNA (1431 ng/µL)
- 1 µL HincII
- 41 µL ddH2O
- 5 µL CutSmart buffer
- * 2h; 37 °C
Results:
generuler; pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2; P74[HincII]
- F1024 49.3 ng\µL
Transformation of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2
Investigator: MT
Aim of the experiment:
- Amplification of Vector
Procedure:
- Transformation following SOP(13.02.2013)
Digestion of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2
Investigator: MT
Aim of the experiment:
- Digestion of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 + PmeI & NotI
Procedure:
- 7 µL DNA (77 ng/µL)
- 0.25 µL PmeI
- 0.25 µL NotI
- 34.5 µL ddH2O
- 5 µL CutSmart buffer
- 2h; 37 °C
Results:
generuler; pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2; P74[HincII]
- F1023 7.6 ng\µL
Gelextraction PCR pSB1C3
Investigator: MT
Aim of the experiment:
- Purification of PCR linear pSB1C3
Procedure:
- According to Gelextraction kit; Quiagen
Results:
generuler; PCR gsy8; PCR linear pSB1C3
- F1025 9.2 ng\µL
Analytical digestion of P249, P268 and P269 (each by XbaI&HindIII)
Investigator: EU
Aim of the experiment: Analyze construct for eventual preparative digestion
Procedure:
5 µl DNA (except for P249 --> 13 µl) 12µl ddH2O (except for P249 --> 4 µl) 1µl of each enzyme 2µl cut smart buffer
incubation at 37°C for 2 h Gel electrophoresis and gel extraction
Results:
In each digestion only one fragment (the vector) was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder.
Preparative digestion of P272 and P273 (each by XbaI&HindIII)
Investigator: EU
Aim of the experiment: Prepare construct for ligation later on
Procedure:
5 µl DNA 36µl ddH2O 2µl of each enzyme 5µl cut smart buffer
incubation at 37°C for 2 h Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol
Results:
Fragment was expected at about 4 kb. This could be shown comparing with 1kb DNA ladder.
Miniprep of P296
Investigator: EU
Aim of the experiment: Obtain plasmids
Procedure:
- Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN)
- Plasmid was not renamed.
Results:
- Concetrations in "Inventarliste"
Sequencing of P296
Investigator: EU
Aim of the experiment: Sequencing of P296
Procedure:
- Sequencing batch was prepared after manufacturer's protocol:
- 7 µl of plasmid DNA (end concentration 50 - 100ng) in 8µl H2O
- 2 µl sequencing primer (one batch with VF2 and another with VR)
- The different vectors we sequenced received the following barcodes:
- P296 with VF2: FR
- P296 with VR: FR
Results:
Digestion of P1007, P1008 with EcoRI HF, SpeI HF + analytical gel
Investigator: EF
Aim of the experiment: Isolation of pSB1C3 plasmid from p1007) Procedure:
- p1007 digested with:
0,25 µl EcoRI (HF) 0,25 µl SpeI (HF), 5 µl CutSmart Buffer 3 µg plasmid-DNA (P1007) fill up with ddH2O (Vtotal= 50µL)
- 2x 25 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard)
Results: Despite optimised enzyme quantity the insert seems to not have been cut out of the plasmid. One single band at 3kb --> repeat reaction; check cutting sites with genious (*)has p1007 been sequenzed before(?). However the band was extracted from the geled and freezed labeled as "8.1"
- p1008 digested with:
0,25 µl EcoRI (HF) 0,25 µl SpeI (HF), 5 µl CutSmart Buffer 3 µg plasmid-DNA (P1008) fill up with ddH2O (Vtotal= 50µL)
- 2x 25 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard)
Results: Insert was isolated by gel extraction following GelExtraction Kit instructions.
Saturday, Aug 27
PCR GSY8
Investigator: MT
Aim of the experiment:
- Amplification of GSY 8
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (VR) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (D20) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 10 sec |
| 61 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Results:
- negativ two times :(
Mung Bean nuclease (MBN) digestion of F1023
Investigator: MT
Aim of the experiment:
- Get rid of overhangs. Preparation of DNA for blunt ligation
Procedure:
- 18 µL DNA (144 ng)
- 2 µL MBN buffer
- 0.5 µL MBN
- 30 min; 30 °C
- purification by PCR purification kit, Promega
Results:
- F1026 24 ng\µL; must be wrong, because this is more than digested. Endvolume 20 µL
Dephosphorylation of F1024
Investigator: MT
Aim of the experiment:
- Preparation of Vekor for blunt end Ligation
Procedure:
- 13 µL DNA (~50 ng\µL)
- 5 µL Antarctic phosphatase (AP)
- 2 µL AP buffer
- 30 min ; 37 °C
- heat inactivation: 2 min; 80 °C
Results:
- F1029
Ligation: F1025 +F1022 = F1027
Investigator: MT
Aim of the experiment:
- Ligation: F1025 +F1022
Procedure:
- 1 µL F1025 (10 ng\µL)
- 10 µL F1022 (5 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- 6 µL ddH2O
- 60 min; 30°C
Results:
- F1025 +F1022 = F1027
Ligation: F1025 +F1026 = F1028
Investigator: MT
Aim of the experiment:
- Ligation: F1025 +F1026
Procedure:
- 1 µL F1025 (10 ng\µL)
- 20 µL F1026 (24 ng\µL)
- 3 µL buffer
- 1 µL T4 ligase
- 4 µL ddH2O
- 60 min; 37 °C
Results:
- F1025 +F1026 = F1028
Phosphorylation of GSY7; GSY8
Investigator: MT
Aim of the experiment:
- Phosphorylation of GSY7; GSY8
Procedure:
- 5 µL DNA
- 2 µL T4 Ligase buffer
- 1 µL T4 PNK
- 12 µL ddH20
- 30 min; 37 °C
- heat inactivation: 20 min; 65 °C
Results:
- F1030 (GSY7)
- F1031 (GSY8)
Over night liquid culture of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2
Investigator: MT
Aim of the experiment:
- Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Ligation: F1029 + F1030 = F1032; F1029 + F1031 = F1033
Investigator: MT
Aim of the experiment:
- Ligation: F1029 + F1030 = F1032; F1029 + F1031 = F1033
Procedure:
- 5 µL F1029 ( ~ 30 ng) {ERROR!}
- 10 µL F1030/F1031 (~ 25 ng)
- 2 µL buffer
- 1 µL T4 ligase
- 2 µL ddH2O
- 60 min; 37 °C
Results:
- F1029 + F1030 = F1032
- F1029 + F1031 = F1033
- Both negativ, due to miscalculation
Transformation of F1027; F1028; F1032; F1033
Investigator: MT
Aim of the experiment:
- Amplification of Vector
Procedure:
- Transformation following SOP(13.02.2013)
Sunday, Aug 28
PCR GSY8
Investigator: MT
Aim of the experiment:
- Amplification of GSY 8
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (D20) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (VR) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 10 sec |
| 58 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Results:
Over night liquid culture of F1032; F1028
Investigator: MT
Aim of the experiment:
- Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Plasmidpreparation of liquid culture of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2
Investigator: MP
Aim of the experiment:
Purification of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2
Procedure:
- According to Mini-prep. kit, SLG
Results:'
- P1009 608.6 ng/µL
Week 16, Aug 29nd - Sep 4th
Monday, Aug 29nd
Plasmid preparation of liquid culture of F1032(clon 1); F1032(clon 2)
Investigator: JL
Aim of the experiment:
Purification of F1032(clon 1); F1032(clon 2)
Procedure:
- According to Mini-prep. kit, Quiagen
Results:
- P 1010 448.4 ng/µL
- P 1011 344.6 ng/µL
Analytical digestion of P1010; P1011
Investigator: JL
Aim of the experiment:
- verify constructs: P1010; P1011
Procedure:
- 2 µL DNA
- 0.5 µL EcoRI
- 0.5 µL PstI
- 1 µL CutSmart buffer
- 6 µL ddH20
Results:
- 1kb ladder; P1010; P1011
PCR and purification of GSY7_new and GSY8
Investigator: EU
Aim of the experiment:
- Amplification of GSY 7_new and GSY 8
Procedure:
Same procedure for both GSYs:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (VR) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (D20) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 10 sec |
| 98 °C | 30 sec | |
| 61 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Purification according to Promega PCR purification kit
Results: GSY7: 9,4 ng/µl GSY8: 1,5 ng/µl, probably contaminated with GSY7_new
Phosphorylation of GSY7_new and GSY8
Investigator: EU
Aim of the experiment:
- Phosphorylation of GSY7_new; GSY8
Procedure:
- 5 µL DNA
- 2 µL T4 Ligase buffer
- 1 µL T4 PNK
- 12 µL ddH20
- 30 min; 37 °C
- heat inactivation: 10 min; 75 °C
Results:
- F1034 (GSY7_new)
- F1035 (GSY8)
Ligation: F1034 + F1029 = F1036; F1035 + F1029 = F1037
Investigator: EU
Aim of the experiment:
- Ligation: F1034 + F1029 = F1036; F1035 + F1029 = F1037
Procedure:
- 5 µL F1029 ( ~ 10 ng)
- 10 µL F1034/F1035 (~ 25 ng)
- 2 µL buffer
- 1 µL T4 ligase
- 2 µL ddH2O
- 60 min; 37 °C
Results:
- F1029 + F1034 = F1036
- F1029 + F1035 = F1037
Preparative digestion of P297, P299, P279, P285, P285, P277, P312 & P251
Investigator: Clemens
Aim of the experiment:
Preparation for cloning in pSB1C3/pASK111
Procedure:
- * Batch for preparative digestion of P297 with AgeI-HF & PstI-HF
| volume | reagent |
| 5 µl | Plasmid DNA P297 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | AgeI HF |
| 1 µl | PstI-HF |
| 220 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P299 with NgOMIV and PstI-HF
| volume | reagent |
| 6 µl | Plasmid DNA P299 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 19 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P279 with NgOMIV & PstI-HF
| volume | reagent |
| 20 µl | Plasmid DNA P279 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 5 µl | ddH2O |
| =30 µl | TOTAL |
- * Batch for preparative digestion of P285 with AgeI-HF & PstI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P297 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | AgeI HF |
| 1 µl | PstI-HF |
| 10 µl | ddH2O |
| =30 µl | TOTAL |
- * Batch for preparative digestion of P277 with AgeI-HF & PstI-HF
| volume | reagent |
| 6 µl | Plasmid DNA P277 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | AgeI HF |
| 1 µl | PstI-HF |
| 19 µl | ddH2O |
| =30 µl | TOTAL |
- * Batch for preparative digestion of P312 with AgeI-HF & PstI-HF
| volume | reagent |
| 5 µl | Plasmid DNA P312 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | AgeI HF |
| 1 µl | PstI-HF |
| 20 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P251 with NgOMIV & PstI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P251 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | NgOMIV |
| 1 µl | PstI-HF |
| 10 µl | ddH2O |
| =30 µl | TOTAL |
- Digestions were incubated for 3h at 37°C
- Fragments were cut from the gel and purified by gelectraction (promega)
Results:
- Fragments were renamed (see Inventarliste) and concentrations were measured
Ligation of F254+ F255, F256+F257, F256+F258 & F260+F312
Investigator: Clemens
Aim of the experiment:
- Ligation
Procedure:
- Batch for Ligation of F254 and F255
| volume | reagent |
| 3 µl | Vector F254 |
| 7 µl | Insert F255 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F257 and F256
| volume | reagent |
| 1.9 µl | Vector F257 |
| 8.1 µl | Insert F256 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F258 and F256
| volume | reagent |
| 1.3 µl | Vector F258 |
| 8.7 µl | Insert F256 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Batch for Ligation of F312 and F260
| volume | reagent |
| 1.2 µl | Vector F312 |
| 8.8 µl | Insert F260 |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | H₂O |
| =20 µl | TOTAL |
- Ligations were performed according to SOP
Analytical digestion and gelelectrophoresis of pASK111, P268 (SUMO-PAS), P6, and P259 (both SA)
Investigator: Niklas
Aim of the experiment: verification before cloning for fermentation
Procedure:
| volume | P268/ P259/ P6/ pASK111 |
| 5,8 µl/ 17 µl/ 8,33 µl/ 14,2 µl | Plasmid DNA |
| 2 µl | cut smart (10x) |
| 0.5 µl | XbaI |
| 0.5 µl | HindIII |
| 11,2 µl/ 0 µl/ 8,67 µl/ 2,8 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
- on of the enzymes was contaminated
Transformation of P310 in BL 21; P6, P259, P268, pASK111, F1034+F1028 in xl1-blue
Investigator: NA
Procedure:
- CaCl2-competent E. coli cells were taken from the stock in –80 °C freezer and gently thawed on ice
- 1 µl (retrafo)/ 7 µl (ligation) of DNA was added to 50 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min heat shock at 37 °C
- Added 0,9 ml LB medium to each tube
- Incubation for 60 min at 37 °C in the 180 rpm cell-culture shaker
- 50 µl of the cell suspension was plated on one plate
- The rest were centrifuged for 1 min at 13,000 rpm and the supernatant was dicarded
- The pellet was resuspended in 50 µl of LB-medium and this concentrated cell suspension was plated on a plate.
Tuesday, Aug 30rd
PCR on GSY7 and GSY8
Investigator: CG
Aim of the experiment:
- Amplification of GSY 7 and GSY 8
Procedure:
Same procedure for both GSYs:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (VR) | 1,5 μl | 0.25 μM |
| 10 μM Reverse Primer (D20) | 1,5 μl | 0.25 μM |
| Template DNA | 2 µL | 10 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 45 cycles | 98 °C | 10 sec |
| 98 °C | 30 sec | |
| 60 °C | 90 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Results:
- GSY8 negative: smear at the upper end of the gel.
Preparative digestion of P270 & P308
Investigator: Clemens
Aim of the experiment:
Preparation for cloning in pSB1C3
Procedure:
- Batch for preparative digestion of P270 with EcoRI-HF & Spe-HF
| volume | reagent |
| 25 µl | Plasmid DNA P270 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF |
| 1 µl | SpeI-HF |
| 0 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P308 with EcoRI-HF and XbaI
| volume | reagent |
| 10 µl | Plasmid DNA P308 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF |
| 1 µl | XbaI |
| 15 µl | ddH2O |
| =30 µl | TOTAL |
- Digestions were incubated for 3h at 37°C
- Fragments were cut from the gel and purified by gelectraction (promega)
Results:
- Fragments were renamed (see Inventarliste) and concentrations were measured
Cloning of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) in pSB1C3
Investigator: CG
Aim of the experiment:
Cloning of pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) in pSB1C3
Procedure:
- 17 µl pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) digestion with 1 µl Mung Bean Nuclease and 2 µl buffer for 30 min. at 30°C to receive blunt ends
- Ligation of 8.7 µl pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 (PmeI+NotI) and 1.3 µl pSB1C3
- Overnight incubation at 16 °C
Retransformation of P258
Investigator: CG
Aim of the experiment:
- Amplification of Vector
Procedure:
- Transformation following SOP(13.02.2013)
Digestion of P1005, P1006, P1007 with XbaI, PstI HF + analytical gel
Investigator: EF
Aim of the experiment: Isolation of insert for further construction of hypoxia system Procedure:
- p1005, p1006, p1007 digested with:
0,25 µl EcoRI (HF) 0,25 µl SpeI (HF), 5 µl CutSmart Buffer 3 µg plasmid-DNA (Pp1005, p1006, p1007) fill up with ddH2O (Vtotal= 50µL)
- 30 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard)
- Digestions were incubated for 1h at 37°C
Results: Insert and vector were clearly separated. All cut vector remained at the expected position of 2kb. Inserts were isolated following extraction kit protocol . F number and concentrations were included into the Inventarliste. Remain of 30µl (with dye) frozen at -20 C -> Labeled as 5.1; 6,1; 7.2
Digestion of P1007 with EcoRI HF, SpeI HF + analytical gel
Investigator: EF
Aim of the experiment: Isolation of vector. Verification of digestion performed on 29th, Friday Procedure:
- p1007 digested with:
0,25 µl EcoRI (HF) 0,25 µl SpeI (HF), 5 µl CutSmart Buffer 3 µg plasmid-DNA (p1007) fill up with ddH2O (Vtotal= 50µL)
- Digestion was incubated for 1h at 37°C
- 30 µl on 1% agarose gel for electrophoresis (2log DNA-ladder as standard)
Results: Again plasmid wasn´t cut. One single band at 3kb. Further procedur: sequence plasmid (?) repeat ligation to generate p1007. Isolate pSB1C3 from another plasmid.
[File to be uploaded]
Ligation of F1001 and F1002
Investigator: EF
Aim of the experiment: generate p1007 due to gel results (since 28th, Thursday) Procedure: Ligation was performed as follows: 6,3 µl F1001, 5,3 µl F1002, 2 µl T4-DNA-Ligase Buffer, 1 µl T4-DNA-Ligase, filled up with ddH20 to 20 µl *3:1 vector-insert ratio
Ligation of F248 and 1015
Investigator: EF 'Aim of the experiment: hypoxia Procedure: Ligation was performed as follows: 1 µl F248, 5 µl F1015, 2 µl T4-DNA-Ligase Buffer, 1 µl T4-DNA-Ligase, filled up with ddH20 to 20 µl *Approximate 3:1 vector insert ratio
Results: Plates incubating over night at 37C
Sequencing of P259 (SA from GSY6; P303 and P304 (eMA)
Investigator: NA
Procedure:
- Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer) - all samples: VF2
- barcodes in stock list
Picking of of E. coli XL1 blue with P6, pASK111, P268
Investigator: NA
Procedure:
- 2 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Inocculation of preculture (P310 eMA-HisTag)
Investigator: NA
Procedure:
- 2 colonies were picked (20 mL LB with Cam)
- the preculture was transferred in a cell culture shaker at 37 °C and were incubated overnight
harvesting SUMO Protease
Investigator: NA
Aim of the experiment: SUMO Protease production
Procedure:
- 6 x 2 L cultures
- centrifuge cultures for 30 min at 6000 rpm
- freeze pellet (120.5 g)
Wednesday, Aug 31st
Over night liquid cultures of P1007 and P1012
Investigator: EF
Aim of the experiment:
Obtaining enough cellmass for plasmid preparation. Liquid cultures of Turbo and XL1 blue cells.
Procedure:
- Inoculation of 5 mL LB-medium with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Phosphorylation of F1036
Investigator: MT
Aim of the experiment:
- Phosphorylation of F1036
Procedure:
- 36 µL DNA (*350 ng)
- 4 µL T4 ligase buffer
- 1 µL PNK
- 30 min; 37 °C
- heat deactivation: 20 min; 65 °C
Ligation: F1029 + F1037 = F1038
Investigator: MT
Aim of the experiment:
Ligation: F1029 + F1037 = F1038
Procedure:
- 0.5 µL F1029 (30 ng\µL)
- 10 µL F1037 (9 ng\µL)
- 2 µL T4 ligase buffer
- 1 µL T4 ligase
- 7 µL ddH2O
- over night ; 16 °C
Results:
- F1029 + F1037 = F1038
Miniprep of various plasmids, analytical digestion, sequencing
Investigator: CG
Aim of the experiment: Obtain plasmids for: eMA, PAS-Lys-Sumo, pASK111, Streptavidin (non-codon-optimized)
Procedure:
- Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN)
- Digestion of about 250 ng for each eMA plasmid with EcoRI and PstI
Results:
- Names and concetrations in "Inventarliste"
- Sequencing results
- Gel:
1. GSY8, 2. - 5.: P319-P322 with EcoRI / PstI
Cloning of Streptavidin (non-codon-optimized) into pASK111
Investigator: JL, NA, CG
Aim of the experiment: Obtain Streptavidin (non-codon-optimized) in expression vector
Procedure:
- Preparative digestion of 42 µl P6 (Streptavidin) and 42 µl P315, 5 µl CutSmart and 1.5 µl XbaI/HindIII
- Gel extraction
- Ligation: 5.9 µl pASK111 + 4.1 µl SA + 2 µl ligase buffer + 7 µl ddH2o + 1 µl T4 ligase
- Transformation according to SOP
Results:
- Gel:
1. Streptavidin (non-codon-optimized), 2. pASK111
Preparative digestion of P296 and P73
Investigator: EU
Aim of the experiment: Prepare construct for ligation later on
Procedure:
5 µl DNA 36µl ddH2O 2µl of each enzyme (P296 --> EcoRI&SpeI, P73 --> EcoRI&XbaI) 5µl cut smart buffer
incubation at 37°C for 3 h Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol
Results:
Fragments were expected at about 1 kb (P296) and 2,5 kb (P73). This could be shown comparing with 1kb DNA ladder.
Ligation: F269 + F270 = F271
Investigator: EU
Aim of the experiment:
- Ligation: F269 + F270 = F271
Procedure:
- 5 µL F269 ( ~ ng)
- 10 µL F270 (~ ng)
- 2 µL buffer
- 1 µL T4 ligase
- 2 µL ddH2O
- ONT; 16 °C
Results:
- F269 + F270 = F271
PCR and purification of GSY7_new and GSY8
Investigator: CG, EU
Aim of the experiment:
- Amplification of GSY 7_new and GSY 8
Procedure:
Same procedure for both GSYs:
- PCR using PfuUltra II Fusion HotStart DNA Polymerase according to manufacturer’s online protocol
| Component | 10 µL reaction | Final concentration |
| PfuUltra II Hotstart PCR Master Mix | 25 µL | 1 X |
| 10 μM Forward Primer (O-038) | 2,5 μl | 0.5 μM |
| 10 μM Reverse Primer (O-039) | 2,5 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 2 min |
| 40 cycles | 98 °C | 10 sec |
| 98 °C | 20 sec | |
| 45-64,7 °C (gradient) | 20 sec | |
| Final extension | 72 °C | 30 sec |
| Hold | 4 °C |
Purification according to Promega PCR purification kit
Results: no amplification recognizable
harvesting SUMO Protease
Investigator: NA
Aim of the experiment: SUMO Protease production
Procedure:
- resuspend pellet in PBS + 10 mM EDTA
- homogenize cells with PANDA
- centrifuge cultures for 30 min at 8000 rpm (protein in supernantant)
- dialysis against IMAC buffer
Inocculation and induction of eMA-HisTag (P310)
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- 2 L LB shaking flask
- the cell culture was incubated at 37 °C
- induction with 200 µl anhydro tetracycliine (2mg/ml) at OD= 0,55
- production over night
Thursday, Sep 1st
Miniprep of F261, F262, F263, F264, I739001, P305 & P258 ,analytical digestion and sequencing
Investigator: Clemens
Aim of the experiment: Confirm sucessful cloning
Procedure:
- Miniprep was performed according to manufacturer's protocol (QIAprep, Qiagen)
- Analytic digestion was performed to check if the cloning was successful
- Batch for analytical digestion of P323/P324 with PstI-HF and EcoRI-HF
| volume | reagent |
| 1 µl/ 0.5 µl | Plasmid DNA P323/P324 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P325/P326 with PstI-HF and EcoRI-HF
| volume | reagent |
| 1 µl/ 0.5 µl | Plasmid DNA P326/P325 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P327/P328 with PstI-HF and EcoRI-HF
| volume | reagent |
| 1 µl/ 0.5 µl | Plasmid DNA P328/P327 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P329/P330 with PstI-HF and EcoRI-HF
| volume | reagent |
| 0.5 µl/ 0.5 µl | Plasmid DNA P329/P330 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7.5 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P331/P332 with PstI-HF and EcoRI-HF
| volume | reagent |
| 0.5 µl/ 0.5 µl | Plasmid DNA P331/P332 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7.5 µl | ddH2O |
| =10 µl | TOTAL |
- Batch for analytical digestion of P333/P334/P322 with AgeI-HF and EcoRI-HF
| volume | reagent |
| 0.5 µl/ 0.5 µl/ 0.5 µl | Plasmid DNA P333/P334/P322 |
| 1 µl | Cutsmart buffer (10x) |
| 0.5 µl | EcoRI-HF (10 U/µl) |
| 0.5 µl | PstI-HF (20 U/µl) |
| 7.5 µl/ 7.5 µl/ 7.5 µl | ddH2O |
| =10 µl | TOTAL |
- Digestions were incubated for 1.5h at 37°C
- Samples were run on an 1% agarose gel for 45 min at 90V
Results:
- Except for P329, P330 and P322 all fragments were of the expected size
- Cloning of the inserts of P29 and P30 was started again.
For confirmation of the Sequence the following plasmids were send for sequencing:
- P323 (VF2 & VR)
- P331 (VF2)
- P333 (VF2)
- P328 (VF2)
- P326 (VF2)
Preparative digestion of P333, P308, P331 and P301
Investigator: Clemens
Aim of the experiment:
- Cloning of new constructs
Procedure:
- Batch for preparative digestion of P333 with NgOMIV and PstI-HF
| volume | reagent |
| 6 µl | Plasmid DNA P333 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | NgOMIV |
| 19 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P333 with AgeI-HF and PstI-HF
| volume | reagent |
| 6 µl | Plasmid DNA P333 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | AgeI-HF |
| 19 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P308 with XbaI and EcoRI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P308 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | XbaI |
| 1 µl | EcoRI-HF |
| 10 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P333 with SpeI-HF and EcoRI-HF
| volume | reagent |
| 8 µl | Plasmid DNA P331 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | EcoRI-HF |
| 1 µl | SpeI-HF |
| 17 µl | ddH2O |
| =30 µl | TOTAL |
- Batch for preparative digestion of P301 with PstI-HF and NgOMIV
| volume | reagent |
| 6 µl | Plasmid DNA P301 |
| 3 µl | Cutsmart buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | NgOMIV |
| 19 µl | ddH2O |
| =30 µl | TOTAL |
- Digestions were incubated for 3h at 37°C
- Samples were applied on a 1% agarose gel and run at 90V for 45min
Results:
- Digestions of P308, P331 and P301 showed the expected fragment sizes.
- These fragments were cut from the gel and purified by gelextraction (Promega)
- Samples were renamed and given Fragmentnumbers
- The Digestions of P333 showed fragments of unexpected length. The Ligation of eMA in pSB1C3 (previous step) failed. This ligation has to be repeated.
PCR and purification of GSY7_new and GSY8
Investigator: MP, EU
Aim of the experiment:
- Amplification of GSY 7_new and GSY 8
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
- each GSY was amplified two times; once with Q5 GC-Enhancer and once without
| Component | 25 µL reaction | Final concentration |
| 5X Q5 Reaction Buffer | 5 µL | 1 X |
| 10 mM dNTPs | 0.5 µL | 200 μM |
| 10 μM Forward Primer (D20) | 1.25 μl | 0.5 μM |
| 10 μM Reverse Primer (VR) | 1.25 μl | 0.5 μM |
| Template DNA | 1 µL | 10 ng |
| Q5 High-Fidelity DNA Polymerase | 0.25 μl | 0.02 U/μl |
| Nuclease-Free Water | to 25 μl (minus 5µl for GC-Enhancer respectively) |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 5 - 10 sec |
| 61 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 16 °C |
Note: Use of the NEB T m Calculator is highly recommended.
Purification according to Promega PCR purification kit
Results: no amplification GSY8 recognizable, GSY7did work
Transformation of F271
Investigator: EU
Aim of the experiment:
- Transformation of F271 for further application
Procedure:
- 3 µL DNA were added to blr cells
- incubation on ice for 20 min
- heatshock in 42°C for 40s
- incubation on ice for 4 min
- 700µl LB-medium were added
- incubation for 1h in 37°C shaker
- centrifugation for 3 min
Results:
Transformation of F1038
Investigator: EU
Aim of the experiment:
- Transformation of F1038 for further application
Procedure:
- 3 µL DNA were added to blr cells
- incubation on ice for 20 min
- heatshock in 42°C for 40s
- incubation on ice for 4 min
- 700µl LB-medium were added
- incubation for 1h in 37°C shaker
- centrifugation for 3 min
Results:
harvesting of eMA-HisTag
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- centrifugate at 6000 rpm for 30 min (OD= 6.15)
- 11,1 g pellet
- SDS-PAGE after protocol
- resuspend pellet in 20 mM Tris/HCl + 50 mM NaCl
- homogenize cells with PANDA
- centrifuge 30 min at 18000 rpm
- denaturate inclusion bodies in 16 ml Gnd-Chl (pH 1.5) over night
results:
marker, eMA before induction, eMA after induction, ULP after induction (SUMO protease)
Friday, Sep 2nd
Ligation of F274+F275, F276+F260 and F279+F240
Investigator: Clemens
Aim of the experiment:
- Cloning of new constructs
Procedure:
Results:
harvesting of eMA-HisTag
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- add 4 ml Gnd-Chl because pellet did not dissolve over night, continue shaking for few hours
- centrifugate for 15 min at 15000 rpm (protein in supernatant)
- refolding in 1.5 L cold PBS over night
purification of SUMO protease using ÄKTA purifier
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- centrifugation
- destroying DNA using ultrasonic for 10 min
- sterilfiltration
- purification due to HisTag affinity of protein to nickel in column (buffer: 20 mM Tris/HCl + 500 mM NaCl
- over night
Procedure:
- purification stopped because of pressure alarm, was restarted an showed no SUMO protease in the sample
Cloning of extended MCS into pDSG and pcDNA5 vector
Investigator: CG
Aim of the experiment: Extending the MCS of pDSG and pcDNA5 for cloning of BioBricks
Procedure:
- Digestion of 5 µl P291 (pDSG) and 15 µl P339 (pcDNA) with SphI and NruI-HF for pDSG and MluI-HF for pcDNA5
- Gelextraction of both signals at about 4 kb
- Oligo-hybridzation of O35/O36 and O36/O37 wo prior phosphorylation to obtain a total concentration of 17 ng/µl
- Ligation of oligo-hybridzation and vector-fragements according to calculations in the Inventarliste and overnight incubation at 16°C
Procedure:
- expected signals after digestion: pDSG with 4 kb and 1.7 kb (lane 1), pcDNA with 4 kb and 1 kb (lane 2)
Miniprep of 2x pcDNA5 vector and 2x Streptavidin (nco) in pASK111
Investigator: CG
Aim of the experiment: Obtain plasmids for pcDNA5 vector and 2x Streptavidin (nco) in pASK111
Procedure:
- Miniprep was performed according to manufacturers protocol (QiaPrep, QIAGEN)
Saturday, Sep 3nd
Transformation of F285; F286
Investigator: MT
Aim of the experiment:
Amplification of F285; F286
Procedure:
- Transformation following SOP(13.02.2013)
Digestion of P1009 + PmeI&NotI
Investigator: MT
Aim of the experiment:
- Digestion of P1009 + PmeI&NotI
Procedure:
- 33 µL DNA (~600 ng/µL)
- 2 µL PmeI
- 1 µL NotI
- 9 µL ddH2O
- 5 µL CutSmart buffer
- incubation for 1h at 37 °C
Results:
- F1039 80 ng/µL
Sunday, Sep 4th
Phosphorylation of F1040 (PCR product GSY7)
Investigator: MT
Aim of the experiment:
- Phosphorylation of F1040 (PCR product GSY7)
Procedure:
- 15 µL F1040 (20 ng/µL)
- 5 µL T4 Ligase buffer
- 1 µL T4 PNK
- 29 µL ddH20
- 30 min; 37 °C
- heat inactivation: 20 min; 65 °C
Results:
- F1041 (PCR product GSY7)
Mung Bean nuclease (MBN) digestion of F1039 (pAAV-CWS_MmZip14_EMCV-IRES_mTagBFP2 [PmeI+NotI])
Investigator: MT
Aim of the experiment:
- Get rid of overhangs. Preparation of DNA for blunt ligation
Procedure:
- 18 µL F1039 ( 80 ng/µL)
- 2 µL 10x CutSmart buffer
- 0.25 µL MBN (10 u/µL)
- 60 min; 30 °C
- purification by PCR purification kit, Promega
Results:
- F1042 30 ng\µL
Ligation: F1042 +F1025 = F1043
Investigator: MT
Aim of the experiment:
- Ligation: F1042 +F1025
Procedure:
- 5.5 µL F1025 (9.2 ng\µL)
- 8.2 µL F1042 (35 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- 3.5 µL ddH2O
- over night; 16 °C
Results:
- F1042 +F1025 = F1043
Ligation: F1041 +F1029 = F1044
Investigator: MT
Aim of the experiment:
- Ligation: F1041 +F1029
Procedure:
- 1.6 µL F1029 (30 ng\µL)
- 23 µL F1041 (6 ng\µL)
- 3 µL buffer
- 1 µL T4 ligase
- 1.3 µL ddH2O
- over night; 16 °C
Results:
- F1041 +F1029 = F1044
Cloning of SUMO-PAS in pASK75
Investigator: JL, EF
Aim of the experiment: Ligation of F158 and "P310" (JL) (SUMO+PAS-Lysin in pASK75):
- 5,6 µL
- 2,4 µL
- 2 µL
- 1 µL T4-DNA-Ligase
- fill up with ddH2O to 20 µL
Transformation in NEB Turbo cells of 10 µL ligation
Week 17, Sep 5th - Sep 11th
Monday, Sep 5th
harvesting of eMA-HisTag
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- centrifugate after refolding at 11500 rpm for 30 min
results:
marker, eMA before induction, after induction, after denaturation and refolding
Inocculation of P341 (E.coli Bl-21; 50 ml LB) in M9 medium for fermentation
Investigator: NA
Procedure:
- M9 medium with Cam (1:1000) was inocculated with preculture (~50 mL) of P258 (pASK111+Streptavidin)
- the culture was incubated in the shaker over night at 37°C
results:
- a 50 ml culture with Ecoli Bl21 was induced
the SDS PAGE shows no streptavidin expression after induction
- next fermentation in pSA (not pASK111)
Tuesday, Sep 6th
Phosphorylation of F1005, F1016
Investigator: EF
Aim of the experiment: to be completed
Ligation of F1045, F1046
Investigator: EF
Aim of the experiment: to be completed
Ligation of F1005, F10016 (GelEx GSY8) in F1013
Wednesday, Sep 7th
harvesting of eMA-HisTag
Investigator: NA
Aim of the experiment: eMA-HisTag production
Procedure:
- precipitation with ammonium sulfate gradient (10% to 90%)
- the percentage was increased in 10% steps, after each addition of ammonium sulfate the solution was centrifugated at 8500 rpm for 20 min
- SDS-PAGE: samples of the supernatants
results:
The pellets of the precipitations with 50%, 60% and 70% were pooled and dialysed against IMAC buffer Next step: Äkta
MidiPrep of pSB1C3 with RFP generator (P4)
Investigator: NA
Aim of the experiment: purification of CMV Nanoluc StrepTag PolyA (P312)
Procedure: MidiPrep was performed according to manufacturer's protocol (QIAprep MidiPrep, Qiagen)
Results:
3268 ng/µl in 80 µl
Inocculation of fermenter
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- inocculation with 50 ml preculture (P20 in LB medium)
- pO2= 60 %; Temp= 37°C, pH~7
- growth over night
- OD next morning: 0.4
Thursday, Sep 8th
Fermentation
Investigator: NA
Aim of the experiment: Streptavidin production
Procedure:
- exponential feed (Amp) for ~13 h (about 3-4 L); start at OD= 7.5 (glucose test negative)
- Induction of streptavidin expression (IPTG) at OD= 30
- production over night
Friday, Sep 9th
harvesting streptavidin
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- OD= 41.4
- centrifuge culture for 1 h at 4800 rpm
- resuspend pellet (275 g)in 20mM Tris HCl + 500 mM NaCl(pH 8)
- homogenize with PANDA
- the resulting lysate was transferred into centrifuge tubes and spun down (4°C, 9000 rpm, 30 mins). The supernatant was cast away and the pellet was resuspended in 6M Gua-HCl (pH = 1.5) at 4°C overnight.
Saturday, Sep 10th
Sunday, Sep 11th
harvesting streptavidin
Investigator: NA
Aim of the experiment: streptavidin production
Procedure:
- the denaturated protein solution was refolded in 15 L 1x PBS (added drop wise) over night
- the solution was centrifugated at 9500 rpm for 20 minutes, the pellet was stored in the freezer, the supernatant will be concentrated via cross flow and precipitated with ammonium sulfate afterwards
results:
Week 18, Sep 12th - Sep 18th
Tuesday, Sep 13th
Digestion of P74
Investigator: MT
Aim of the experiment:
- Digestion of P74
Procedure:
F1055:
- 2 µL DNA (1431 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL HincII (10 u/µL)
- 16 µL ddH2O
- incubation for 1h at 37 °C
F1056:
- 2 µL DNA (1431 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL HincII (10 u/µL)
- 0.25 µL AgeI (20 u/µL)
- 15.5 µL ddH2O
- incubation for 1h at 37 °C
F1057:
- 2 µL DNA (1431 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL HincII (10 u/µL)
- 0.25 µL NgoMIV
- 15.5 µL ddH2O
- incubation for 1h at 37 °C
Results:
1Kb ruler, F1055[HincII], F105[HincII&AgeI]6, F1057[HincII&NgoMIV], P1017[EcoRI&SpeI]
- F1055 49.5 ng/µL
- F1056 37.0 ng/µL
- F1057 29.7 ng/µL
Analytical digestion of P1017
Investigator: MT
Aim of the experiment:
- Analytical digestion of P1017
Procedure:
- 2 µL DNA (312 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL EcoRI
- 15.5 µL ddH2O
- incubation for 1h at 37 °C
Results:
1Kb ruler, F1055[HincII], F105[HincII&AgeI]6, F1057[HincII&NgoMIV], P1017[EcoRI&SpeI]
PCR of GSY10
Investigator: MT
Aim of the experiment:
Amplification of GSY10
Procedure:
- PCR Using Q5® High-Fidelity DNA Polymerase (M0491) according to online protocol
| Component | 50 µL reaction | Final concentration |
| Q5® High-Fidelity 2X Master Mix | 25 µL | 1 X |
| 10 μM VR | 2,5 μl | 0.5 μM |
| 10 μM VF2 | 2,5 μl | 0.5 μM |
| Template DNA | 0.1 µL | 1 ng |
| Nuclease-Free Water | to 50 μl |
Thermocycling Conditions for a Routine PCR:
| STEP | TEMP | TIME |
| Initial Denaturation | 98 °C | 30 sec |
| 35 cycles | 98 °C | 10 sec |
| 62 °C | 30 sec | |
| 72 °C | 60 sec | |
| Final extension | 72 °C | 120 sec |
| Hold | 4 °C |
Dephosphorylation of F1055
Investigator: MT
Aim of the experiment:
- Preparation of Vekor for blunt end Ligation
Procedure:
- 15 µL F1055 (~50 ng\µL)
- 1 µL FastAP (1 u/µL)
- 2 µL 10x FastAP buffer
- 10 min ; 37 °C
- heat inactivation: 5 min; 75 °C
Results:
- F348
Digestion of P1017
Investigator: MT
Aim of the experiment:
- Digestion of P1017
Procedure:
- 10 µL DNA (312 ng/µL)
- 5 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL PstI
- 34.5 µL ddH2O
- incubation for 1h at 37 °C
Results:
1Kb ruler, P1017[SpeI&PstI], P1017[SpeI&PstI], F362: PCR-GSY7[AgeI], PCR-GSY7, PCR-GSY8
- F350
Over night liquid culture of P1009
Investigator: MT
Aim of the experiment:
- Getting enough cellmass for plasmid preparation
Procedure:
- Inoculation of 5 mL LB-medium (containing antibiotica) with colonies of foregone transformation
- Incubate over night at 37 °C in shaker
Wednesday, Sep 14th
Gelextraction of F357[NgoMIV]; P1006[XbaI&PstI]
Investigator: MT
Aim of the experiment:
- Gelextaction of F357[NgoMIV]; P1006[XbaI&PstI]
Procedure:
- Using gelextraction kit, Promega
Results:
1kb ladder, F357[NgoMIV]; P1006[XbaI&PstI]
- F357[NgoMIV] -> F364 41 ng/µL
- P1006[XbaI&PstI] -> F365 9.1 ng/µL
Ligation: F1057 + F364 = F366
Investigator: MT
Aim of the experiment:
- Ligation: F1057 + F364
Procedure:
- 1,6 µL F1057 (29.7 ng\µL)
- 2.1 µL F364 (41 ng\µL)
- 3 µL buffer
- 1 µL T4 ligase
- 22.3 µL ddH2O
- 30 min; RT
Results:
- F1057 + F364 = F366
Ligation: F350 + F365 = F367
Investigator: MT
Aim of the experiment:
- Ligation: F350 + F365
Procedure:
- 2,6 µL F350 (19 ng\µL)
- 2.1 µL F365 (9.1 ng\µL)
- 3 µL buffer
- 1 µL T4 ligase
- 15.4 µL ddH2O
- 30 min; RT
Results:
- F350 + F365 = F367
Digestion of P1008
Investigator: MT
Aim of the experiment:
- Digestion of P1008
Procedure:
- 10 µL P1008 (256.5 ng/µL)
- 5 µL CutSmart buffer
- 0.25 µL XbaI
- 0.25 µL PstI
- 34.5 µL ddH2O
- incubation for 1h at 37 °C
Results:
1Kb ruler, ... P1008[XbaI&PstI]
- P1008 -> F368 ~1 ng/µL
- Maybe do it again, fragments do not have the right size, apparently.
Transformation of F366; F367
Investigator: MT
Aim of the experiment:
- Amplification of Vector
Procedure:
- Transformation following SOP(13.02.2013)
Analytical digestion of P296 and F269
Investigator: EU
Aim of the experiment: Analyze construct for eventual preparative digestion
Procedure:
2 µl DNA 15µl ddH2O 1µl of PstI 2µl cut smart buffer
incubation at 37°C over night Gel electrophoresis and gel extraction
Results:
Preparative digestion of P313, P296 and P73
Investigator: EU
Aim of the experiment: Prepare construct for ligation later on
Procedure:
10 µl (P73)/ 15 µl (P296)/ 20 µl (P313) DNA 2µl of each enzyme (P296&P313 --> EcoRI&SpeI, P73 --> EcoRI&XbaI) 5µl cut smart buffer ad 50 µl ddH2O
incubation at 37°C over night Gel electrophoresis and gel extraction by Gel extraction kit (Promega) according to manufacturer’s protocol
Results:
- P296 [EcoRI&SpeI] --> F378
- P313 [EcoRI&SpeI] --> F383
- P73 [EcoRI&XbaI] --> F379
Ligation: F269 + F270 = F380; F269 + F92 = F381; F269 + F133 = F382
Investigator: EU
Aim of the experiment:
- Ligation:
- F269 + F270 = F380
- F269 + F92 = F381
- F269 + F133 = F382
Procedure:
- 3 µL F269 (6,3 ng)
- 0,5 µL F270 (5,6ng)/ 0,25 µl F92 (6,75 ng)/ 0,25 µl F133 (13 ng)
- 0,5 µL buffer
- 0,5 µL T4 ligase
- 0,5/0,75 µL ddH2O
- ONT; 16 °C
Results:
- F269 + F270 = F380
- F269 + F92 = F381
- F269 + F133 = F382
Sequencing of P313
Investigator: EU
Aim of the experiment: Sequencing of P313
Procedure:
- Sequencing batch was prepared after manufacturer's protocol:
- 6 µl of plasmid DNA (end concentration 58,6 ng) in 9µl H2O
- 2 µl sequencing primer (VF2)
- The different vectors we sequenced received the following barcodes:
- P313 with VF2: FR12904387
Results:
Thursday, Sep 15th
Digestion of P1008
Investigator: MT
Aim of the experiment:
- Digestion of P1008
Procedure:
- 10 µL P1008 (256.5 ng/µL)
- 5 µL CutSmart buffer
- 0.25 µL XbaI
- 0.25 µL PstI
- 34.5 µL ddH2O
- incubation for over night at 37 °C
Results:
P1008[XbaI&PstI], P1008[XbaI&PstI], 1Kb ruler
- P1008[XbaI&PstI] -> F375 32 ng/µL
Ligation: F63 + F375 = F377
Investigator: MT
Aim of the experiment:
- Ligation: F63 + F375
Procedure:
- 2.2 µL F63 (45 ng\µL)
- 6.3 µL F375 (32 ng\µL)
- 3 µL buffer
- 1 µL T4 ligase
- 17.5 µL ddH2O
- 30 min; RT
Results:
- F63 + F375 = F377
Fill in of overhangs of F386 with Large Klenow fragment
Investigator: MT
Aim of the experiment:
- 3' overhang removal and fill-in of 3' recessed (5' overhang) ends using DNA Polymerase I, Large (Klenow) Fragment
- short: create blunt ends, needed for ligation
Procedure:
- 17.09 µL F386 (25.9 µL)
- 0.66 µL 10 mM dNTP Mix
- 2 µL T4 Ligase buffer
- 0.25 µL Klenow Fragment
- 15 min; RT
- heat inactivation: 20 min; 75 °C
Results:
- F387
Incubation of F386 with Q5 polymerase
Investigator: MT
Aim of the experiment:
- Create blunt ends, needed for ligation
Procedure:
- 5 µL 2x Q5 Mastermix
- 5 µL F386 (25.9 ng/µL)
- 15 min; 72 °C
Results:
- F388
Ligation: F374 + F386 = F390
Investigator: MT
Aim of the experiment:
- Ligation: F374 + F386
Procedure:
- 1 µL F374 (25.9 ng\µL)
- 17.5 µL F387 (~ 22 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- 30 min; RT
Results:
- F374 + F386 = F390
Ligation: F374 + F388 = F391
Investigator: MT
Aim of the experiment:
- Ligation: F374 + F388 = F391
Procedure:
- 0.3 µL F374 (25.9 ng\µL)
- 10 µL F388 (~ 13 ng\µL)
- 2 µL T4 ligase buffer
- 1 µL T4 ligase
- 6.7 µL ddH2O
- 30 min; RT
Results:
- F374 + F388 = F391
Transformation of F390, F391, F376 and F377 (INSULIN)
Investigator: EU
Aim of the experiment:
- Transformation of F390, F391, F376 and F377 for further application
Procedure:
- 3 µL DNA were added to xl1-cells
- incubation on ice for 20 min
- heatshock in 42°C for 40s
- incubation on ice for 4 min
- 700µl LB-medium were added
- incubation for 1h in 37°C shaker
- centrifugation for 3 min
Results:
Transformation of F389 (zk-vector)
Investigator: EU
Aim of the experiment:
- Transformation of F389 for further application
Procedure:
- 3 µL DNA were added to xl1-cells
- incubation on ice for 20 min
- heatshock in 42°C for 40s
- incubation on ice for 4 min
- 700µl LB-medium were added
- incubation for 1h in 37°C shaker
- centrifugation for 3 min
Results:
Transformation of F366
Investigator: EU
Aim of the experiment:
- Transformation of F366 for further application
Procedure:
- 3 µL DNA were added to xl1-cells
- incubation on ice for 20 min
- heatshock in 42°C for 40s
- incubation on ice for 4 min
- 700µl LB-medium were added
- incubation for 1h in 37°C shaker
- centrifugation for 3 min
Results:
Friday, Sep 16th
Digestion of P430
Investigator: MT
Aim of the experiment:
- Digestion of P430
Procedure:
- 10 µL 430 (~150 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL PacI
- 0.25 µL AgeI
- 7.5 µL ddH2O
- 1h; 37 °C
Results:
1kb ruler; P430[PacI&AgeI], P424[SpeI&PstI], P425[SpeI&PstI]
- P430[PacI&AgeI] -> F400 2.1 ng/µL
Digestion of P424
Investigator: MT
Aim of the experiment:
- Digestion of P424
Procedure:
- 17 µL 424 (60.9 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL PstI
- 1h; 37 °C
Results:
1kb ruler; P430[PacI&AgeI], P424[SpeI&PstI], P425[SpeI&PstI]
- P424[SpeI&PstI] -> F401 15.7ng/µL
Week 19, Sep 19th - Sep 25th
Tuesday, Sep 20th
Digestion of P444; P445; P446
Investigator: MT
Aim of the experiment:
- Digestion of P444; P445; P446
Procedure:
P444:
- 8 µL P444 (377.7 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL PstI
- 9.5 µL ddH2O
- 1h; 37 °C
P445:
- 14 µL P445 (223.7 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL PstI
- 3.5 µL ddH2O
- 1h; 37 °C
P446:
- 17.5 µL P446 (144 ng/µL)
- 2 µL CutSmart buffer
- 0.25 µL SpeI
- 0.25 µL PstI
- 1h; 37 °C
Results:
1kb ruler P451[PacI&MluI], P451[PacI&MluI], P444[SpeI&PstI], P446[SpeI&PstI]
-> I did P444 again, because there was marker in the line.
1kb ruler, P444[SpeI&PstI], P445[SpeI&PstI]
- P444[SpeI&PstI] -> F420 18 ng/µL
- P445[SpeI&PstI] -> F418 24.2 ng/µL
- P446[SpeI&PstI] -> F421 7.4 ng/µL
Wednesday, Sep 21th
Ligation {F438 + F435} + F433 or F334
Investigator: MT
Aim of the experiment:
- Ligation {F438 + F435} + F433 or F334
Procedure:
F438 + F435:
- 10 µL F438 (40.2 ng/µL)
- 11 µL F435 (30.5)
- 2 µL T4 Ligase buffer
- 1 µL T4 Ligase
- 6 µL ddH2O
{F438 + F435} + F433:
- 0.5 µL F433 (48.2 ng/µL)
- 4.5 µL {F438 + F435} (~24.4 ng/µL)
- 2 µL T4 Ligase buffer
- 1 µL T4 Ligase
- 12 µL ddH2O
{F438 + F435} + F434:
- 0.5 µL F434 (109 ng/µL)
- 4.5 µL {F438 + F435} (~24.4 ng/µL)
- 2 µL T4 Ligase buffer
- 1 µL T4 Ligase
- 11.5 µL ddH2O
Results:
- F438 + F435 = F439
- {F438 + F435} + F434 = F440
- {F438 + F435} + F433 = F441
Sunday, Sep 25th
Ligation: F457 + F463 = F464
Investigator: MT
Aim of the experiment:
- Ligation: F457 + F463
Procedure:
- 2.7 µL F457 (29.3 ng\µL)
- 4.3 µL F463 (22.1 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- 30 min; RT
Results:
- F457 + F463 = F464
Ligation: F458 + F463 = F465
Investigator: MT
Aim of the experiment:
- Ligation: F458 + F463
Procedure:
- 3.8 µL F458 (15.5 ng\µL)
- 3.2 µL F463 (22.1 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- 30 min; RT
Results:
- F458 + F463 = F465
Ligation: F434 + F464 = F466
Investigator: MT
Aim of the experiment:
- Ligation: F434 + F464
Procedure:
- 0.5 µL F434 (109.1 ng\µL)
- 6.5 µL F464 (17.4 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- over night; 16 °C
Results:
- F434 + F464 = F466
Ligation: F434 + F465 = F467
Investigator: MT
Aim of the experiment:
- Ligation: F434 + F465
Procedure:
- 0.4 µL F434 (109.1 ng\µL)
- 6.6 µL F465 (12.96 ng\µL)
- 2 µL buffer
- 1 µL T4 ligase
- over night; 16 °C
Results:
- F434 + F465 = F467