(→Collaboration with TU Darmstadt iGEM Team) |
(→HowTo) |
||
| (43 intermediate revisions by 8 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{LMU-TUM_Munich|navClass= | + | {{LMU-TUM_Munich|navClass=collaborations}} |
| − | + | <div class="float-right">[[Image:Muc16_Sticker_Collaboration_003.png|350px|link=]]</div> | |
| − | [[ | + | |
=Scientific Collaborations= | =Scientific Collaborations= | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Collaboration with Sheffield iGEM Team== | ==Collaboration with Sheffield iGEM Team== | ||
| − | [[ | + | <div class="white-box"> |
| − | We met the [https://2016.igem.org/Team:Sheffield iGEM Team from Sheffield University] at the European Experience in Paris where we presented our projects to each other. When | + | <div class="imagelink float-right">[[Media:Muc16_Collaboration_Lcn2.png]][[Image:Muc16_Collaboration_Lcn2.png|500px|link=]] |
| − | < | + | <div class="caption"> |
| + | '''Collaboration with [https://2016.igem.org/Team:Sheffield iGEM Sheffield ]: recombinant Lcn2'''<br>'''A)''' Production and purification strategy, '''B)''' Size-exclusion chromatography shows a monodisperse protein, '''C)''' the purified Lcn2 protein is >98% pure under reducing and non-reducing conditions in SDS-PAGE, '''D)''' crystal structure of Lcn2 (PDB: [http://www.rcsb.org/pdb/explore.do?structureId=3i0a| 3I0A]) together with the bound siderophore enterobactin and an iron atom. | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | We met the [https://2016.igem.org/Team:Sheffield iGEM Team from Sheffield University] at the European Experience in Paris where we presented our projects to each other. When they told us that they were using the human [https://en.wikipedia.org/wiki/LCN2 Lipocalin 2 (Lcn2)] in order to detect the siderophores of pathogenic bacteria, we came up with the idea that we could provide the team with highly pure Lcn2 protein. This was a fruitful collaboration as our team's main PI, Prof. Dr. Arne Skerra, is one of the leading experts in the field of lipocalins<ref>Schiefner, A., & Skerra, A. (2015). The menagerie of human lipocalins: a natural protein scaffold for molecular recognition of physiological compounds. Accounts of chemical research, 48(4), 976-985.</ref>. For this reason, the expression constructs and the protocols to produce this recombinant protein are well established our affiliated lab. The main challenge in producing the recombinant protein was that the Sheffield iGEM team required the apo form of the protein without the bacterial siderophore as they aimed to scavenge siderophores from samples and then determine the siderophore content. Thus, we produced the recombinant protein in ''E. coli'' such that it was free from the siderophore generated in conventional ''E. coli'' strains. | ||
| + | </div> | ||
==Collaboration with Erlangen iGEM Team== | ==Collaboration with Erlangen iGEM Team== | ||
| − | [[ | + | <div class="white-box"> |
| − | The iGEM team from [https://2016.igem.org/Team:FAU_Erlangen| FAU Erlangen] | + | <div class="imagelink float-right">[[Media:Muc16_Collaboration_eGFP.png]][[Image:Muc16_Collaboration_eGFP.png|500px|link=]]<div class="caption"> |
| − | < | + | '''Collaboration with [https://2016.igem.org/Team:FAU_Erlangen iGEM FAU Erlangen]: '''<br>'''A)''' Production and purification strategy, '''B)''' Size-exclusion chromatography, '''C)''' Filtration, '''D)''' Tube readied for shipment, '''E)''' Yield after purification |
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | |||
| + | The iGEM team from [https://2016.igem.org/Team:FAU_Erlangen| FAU Erlangen] was working on the development of a biological photovoltaic cell, for which they aimed to use fluorescent proteins to harvest the photons. We saw their presentation at the iGEM Germany meet-up in Marburg, and as we used the purified recombinant eGFP for our own project in any case, we proposed to provide them with eGFP for their experiments. Luckily, at the "Biotechnikum" of the TUM, an annual lab course for bioprocess engineering students produces a large batch of eGFP in ''E. coli'' at a bioprocess scale of several hundred liters of culture medium. For our project, we were provided with ≈50 g of eGFP that we subsequently purified by ammonium sulfate precipitation as well as immobilized metal affinity chromatography (IMAC), and eventually dialyzed the protein against PBS. At the beginning of October, we provided 1.4 g of purified eGFP to the [https://2016.igem.org/Team:FAU_Erlangen| FAU Erlangen] iGEM team. | ||
| + | </div> | ||
==Collaboration with TU Darmstadt iGEM Team== | ==Collaboration with TU Darmstadt iGEM Team== | ||
| − | <div class"white-box">[[ | + | <div class="white-box"> |
| − | In order to | + | <div class="imagelink float-right">[[Media:Muc16_Collaboration_syringe_pump.png]][[Muc16_Collaboration_syringe_pump.png|500px|link=]]<div class="caption"> |
| + | '''Collaboration with [https://2016.igem.org/Team:TU_Darmstadt iGEM FAU Erlangen]: '''<br>'''A)''' , '''B)''' | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | |||
| + | In order to obtain first feedback for our DIY bioprinter kit, the iGEM Team from the [https://2016.igem.org/Team:TU_Darmstadt| TU Darmstadt] agreed to become our beta-tester for the syringe pump that we had constructed for our bioprinter as well as for the whole Synbio community. Our syringe pump was proven to work well in our lab and it can be built with an investment of less than 50 USD provided an off-the-shelf 3D printer is available. For the beta test, the Darmstadt team received only the files for the 3D prints as well as all the parts (such as a stepper motor, few screws, some end-stop switches and the necessary wiring). | ||
| + | |||
| + | Here is their feedback from after they assembled our syringe pump: | ||
| − | + | '''''"'''The pump is neat and stable. The idea of using buttons as end stops is, in principle, good. You took the necessary holes into account and we were able to assemble it according to the instructions. | |
| − | '''''"'''The pump is neat and stable. The idea of using buttons as end stops is in principle good. You took the necessary holes into account and we were able to assemble it according to the instructions. | + | |
''The click system for the syringe was well made and worked straightaway. The linear bearing could be used right away as well. | ''The click system for the syringe was well made and worked straightaway. The linear bearing could be used right away as well. | ||
| − | ''We utilized a homemade 3D printer on highest gear for printing the parts. As you might know this leads to relatively many "aberrations" and errors in the prints. They made the assembly difficult and we had to post process several of them. Your construction is designed quite closed, which renders it difficult for any passionate tinkerer to make adaptations after the printing. For instance we noted that you had sent us a printed clutch though the stepper motor was not shortened and did not fit. | + | ''We utilized a homemade 3D printer on highest gear for printing the parts. As you might know, this leads to relatively many "aberrations" and errors in the prints. They made the assembly difficult and we had to post-process several of them. |
| − | ''In general there are different stepper motors in use in the DIY community | + | <div class="imagelink float-left">[[Media:Muc16_Collaboration_syringe_pump2.png]][[Image:Muc16_Collaboration_syringe_pump2.png|500px|link=]] |
| − | ''Furthermore the suspension device for the syringe could be improved, though the varying design of DIY printers makes finding a "good" solution rather difficult.'''"''''' | + | </div><!-- imagelink --> |
| + | Your construction is designed quite closed, which renders it difficult for any passionate tinkerer to make adaptations after the printing. For instance, we noted that you had sent us a printed clutch though the stepper motor was not shortened and did not fit. Therefore, we drilled a hole in the front, enlarged the clutch and glued the parts together.'' | ||
| + | ''In general, there are different stepper motors in use in the DIY community; hence, we suggest to conceptualize a more general clutch to ease this problem.'' | ||
| + | ''Furthermore, the suspension device for the syringe could be improved, though the varying design of DIY printers makes finding a "good" solution rather difficult.'''"''''' | ||
</div> | </div> | ||
| Line 33: | Line 46: | ||
== The European Experience, Paris July 2nd to 3rd== | == The European Experience, Paris July 2nd to 3rd== | ||
| − | + | <div class="white-box"> | |
| − | Following the invitation by the teams from iGEM [https://2016.igem.org/Team:Ionis_Paris IONIS] and [https://2016.igem.org/Team:Evry Evry] we packed our suitcases and off | + | The first big journey for our team lead us all the way to Paris, France. |
| − | [[ | + | Following the invitation by the teams from iGEM [https://2016.igem.org/Team:Ionis_Paris IONIS] and [https://2016.igem.org/Team:Evry Evry], we packed our suitcases and were off. |
| − | [[ | + | <div class="imagelink float-left">[[Media:Muc16_Paris_1.jpeg]][[Image:Muc16_Paris_1.jpeg|250px|link=]]<div class="caption"> |
| − | And after a little climbing tour to Montmartre and maybe some | + | Munich |
| − | Early | + | </div><!-- caption --> |
| − | which ended | + | </div><!-- imagelink --> |
| + | <div class="imagelink float-left">[[Media:Muc16_Paris_2.jpeg]][[Image:Muc16_Paris_2.jpeg|250px|link=]]<div class="caption"> | ||
| + | Paris | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | <div class="imagelink float-left">[[Media:Muc16_Paris_3.jpeg]][[Image:Muc16_Paris_3.jpeg|250px|link=]]<div class="caption"> | ||
| + | Montmartre | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | <div class="imagelink float-right">[[Media:Muc16_Teamphoto_NA.JPG]][[Image:Muc16_Teamphoto_NA.JPG|300px|link=]]<div class="caption"> | ||
| + | Team stand | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | |||
| + | And after a little climbing tour to Montmartre and maybe some super-last-minute poster printing ;), we settled for the night in our shady "lodging" to be ready to showcase our project publicly for the first time. | ||
| + | Early morning, we headed to the event location. First objective: '''COFFEE''',<br> | ||
| + | which ended up being a greater challenge than could have been anticipated... | ||
Don't bring a knife (''instant coffee'') to a gun fight (''horde of thirsty, tired students'') :D <br> | Don't bring a knife (''instant coffee'') to a gun fight (''horde of thirsty, tired students'') :D <br> | ||
| − | [[ | + | <div class="imagelink float-left">[[Media:Muc16_Paris_4.JPG]][[Image:Muc16_Paris_4.JPG|180px|link=]]<div class="caption"> |
| − | + | Fire performance | |
| − | It was great to see all those ideas and of course to be able to | + | </div><!-- caption --> |
| − | After the poster sessions was over and | + | </div><!-- imagelink --> |
| − | [[ | + | |
| − | The evening promised to be a great dance party in "downtown | + | After breakfast was acquired, things started to get interesting. Dozens of awesome projects from all over Europe and even the UK! |
| − | The midnight surprise | + | It was great to see all those ideas and of course to be able to showcase our own bioprinting project ourselves. We also obtained helpful tips and hints. |
| − | With this awesome party night marking the end of the [http://www.facebook.com/events/458143747717965/ ''European Experience''] we used the following Sunday to enjoy a wonderful | + | After the poster sessions was over and nutrition had been taken care of, interesting panels on ethics and entrepreneurship in synthetic biology followed before it was time for Randy to drop some final Brexit jokes and conclude the scientific section of the event. |
| − | < | + | <div class="imagelink float-right">[[Media:Muc16_Paris_5.JPG]][[Image:Muc16_Paris_5.JPG|300px|link=]]<div class="caption"> |
| + | Party | ||
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | |||
| + | The evening promised to be a great dance party in "downtown Paris," including a live stream of the quarter-final match of the European Championship between Italy and of course Germany. After this sweat-breaking, super-tight, overly nerve-wracking game was finally over, with the better end for us :), we hit the dance floor down in the underground car park. | ||
| + | The midnight surprise: a breathtaking fire-breather, who for some reason was interrupted by the security guard, we wonder why ;). | ||
| + | With this awesome party night marking the end of the [http://www.facebook.com/events/458143747717965/ ''European Experience''], we used the following Sunday to enjoy a wonderful French breakfast and explore the tourist sites of the "City of Love" before it was time to board the plane back to the lab. What a great event! | ||
| + | </div> | ||
== iGEM meets Marburg, Marburg August 5th to 7th== | == iGEM meets Marburg, Marburg August 5th to 7th== | ||
| + | <div class="white-box"> | ||
Just one month after the ''European Experience'' it was time for the next big meetup, the German meetup in the historic city Marburg with its winding streets and steep and tiny alleys. | Just one month after the ''European Experience'' it was time for the next big meetup, the German meetup in the historic city Marburg with its winding streets and steep and tiny alleys. | ||
| − | [[ | + | <div class="imagelink float-left">[[Media:Muc16_Marburg1.JPG]][[Image:Muc16_Marburg1.JPG|400px|link=]] |
| − | Just after arriving at the camp site and setting everything up, the first official point of the program was already | + | </div><!-- imagelink --> |
| − | The next morning the teams from literally all over Germany and extended | + | <div class="imagelink float-right">[[Media:Muc16_Marburg2.JPG]][[Image:Muc16_Marburg2.JPG|400px|link=]] |
| − | After | + | </div><!-- imagelink --> |
| − | + | ||
| + | Instead of boarding a plane, we loaded the car and went on a little road trip of 450 km through wind and rain. Luckily the weather got better and better the closer we came. | ||
| + | Just after arriving at the camp site and setting everything up, the first official point of the program was already about to begin: bar tour, yeah! A long and noisy night followed ;) | ||
| + | The next morning, the teams from literally all over Germany and extended Germany, also known as Denmark, assembled at the main event building and grabbed some awesome breakfast, including appropriate amounts of coffee, before being welcomed by our great hosts from [http://www.facebook.com/igemmarburg iGEM Marburg]. | ||
| + | After introduction speeches by the sponsors of the event, the teams took the stage and presented their projects. As some time had passed since the Paris event, we got to see how the ideas had developed, and there were also some we had not seen before. Really awesome! | ||
| + | After grabbing lunch, it was our turn to show what we had done and would still do in the lab. | ||
<html><video width="300" style="float:left; margin-right : 15px; margin-top : 15px" autoplay muted loop controls> | <html><video width="300" style="float:left; margin-right : 15px; margin-top : 15px" autoplay muted loop controls> | ||
<source src="https://static.igem.org/mediawiki/2016/8/8f/Muc16_Marburg3.mp4" type="video/mp4"> | <source src="https://static.igem.org/mediawiki/2016/8/8f/Muc16_Marburg3.mp4" type="video/mp4"> | ||
| Line 64: | Line 107: | ||
The afternoon was dedicated to physical activity of a special kind, [https://en.wikipedia.org/wiki/Bubble_bump_football '''Bubble Soccer''']. Probably the most hilarious game ever, though it will drain all your energy. Luckily our hosts had anticipated this and provided us with a tasty BBQ. | The afternoon was dedicated to physical activity of a special kind, [https://en.wikipedia.org/wiki/Bubble_bump_football '''Bubble Soccer''']. Probably the most hilarious game ever, though it will drain all your energy. Luckily our hosts had anticipated this and provided us with a tasty BBQ. | ||
Though everyone was a bit tired, the day was far from over as there was the final party of the weekend. | Though everyone was a bit tired, the day was far from over as there was the final party of the weekend. | ||
| − | The next morning came way | + | The next morning came way too quickly, but luckily it was a beautiful sunny morning. We had the chance to hike to the castle and enjoy the amazing view from the top before it was again time to say Goodbye for a while and hit the road back to the lab. |
Thanks again Marburg for having us, it was a pleasure! | Thanks again Marburg for having us, it was a pleasure! | ||
| − | < | + | </div> |
= Helping other iGEM teams = | = Helping other iGEM teams = | ||
==iGEM experiment protocols in all languages by the METU HS Ankara iGEM team== | ==iGEM experiment protocols in all languages by the METU HS Ankara iGEM team== | ||
| − | [[ | + | <div class="white-box"> |
| − | This | + | <div class="imagelink float-right">[[Media:Muc16_Ankara.jpg]][[Image:Muc16_Ankara.jpg|400px|link=]] |
| + | </div><!-- imagelink --> | ||
| + | |||
| + | This year's [https://2016.igem.org/Team:METU_HS_Ankara/Collaborations team of Highschool students from Ankara] got in touch with us and asked for help for their clever idea of translating the standard iGEM lab protocols into different languages. This will hopefully help future teams from all around the globe to better understand them and apply them in the lab without being required to understand English perfectly. Especially other future Highschool teams should profit from this.<br> | ||
We are happy to be part such a great project and make iGEM even more international. | We are happy to be part such a great project and make iGEM even more international. | ||
| − | < | + | </div> |
==65 roses campaign by the iGEM team Tel Hai== | ==65 roses campaign by the iGEM team Tel Hai== | ||
| − | [[ | + | <div class="white-box"> |
| − | As part of their project dealing with the most prevalent mutation in the CFTR protein, responsible for | + | <div class="imagelink float-right">[[Media:Muc16_65roses.png]][[Image:Muc16_65roses.png|400px|link=]]<div class="caption"> |
| − | < | + | A rose from Munich |
| + | </div><!-- caption --> | ||
| + | </div><!-- imagelink --> | ||
| + | |||
| + | As part of their project dealing with the most prevalent mutation in the CFTR protein, which is responsible for cystic fibrosis, the [https://2016.igem.org/Team:Tel-Hai team from Tel Hai Academic college] dedicated themselves to raise awareness for the severely life-shortening genetic condition. The [https://www.cff.org/About-Us/About-the-Cystic-Fibrosis-Foundation/The-65-Roses-Story/ “65 Roses” story] itself dates back to 1965 when an observant 4-year-old, hearing the name of his disease for the first time, pronounced cystic fibrosis as "65 Roses." Nowadays, “65 Roses” is a term often used by young children with cystic fibrosis to pronounce the name of their disease. We are very happy to be a part of their noble efforts and dedication for this project. | ||
| + | </div> | ||
==BioBricks for the world== | ==BioBricks for the world== | ||
| − | Due to the history of interesting projects from Munich it did not take long for the first Biobricks | + | <div class="white-box"> |
| − | [[ | + | Due to the history of interesting projects from Munich, it did not take long for the first Biobricks requests to come in from all over the world. |
| + | <div class="imagelink float-right">[[Media:Muc16_Toulouse.jpg]][[Image:Muc16_Toulouse.jpg|200px|link=]] | ||
| + | </div><!-- imagelink --> | ||
| + | |||
===Vive le France!=== | ===Vive le France!=== | ||
| − | The first inquiry came from our [https://2016.igem.org/Team:Toulouse_France cave saving colleagues from Toulouse] were in need | + | The first inquiry came from our [https://2016.igem.org/Team:Toulouse_France cave-saving colleagues from Toulouse], who were in need of a [http://parts.igem.org/Part:BBa_K823026 BioBrick for their ''Bacillus subtilis''] originally developed by the team from LMU in 2012. Go save those caves! ;) |
===The far east=== | ===The far east=== | ||
| − | Not long after a more distant call reached us all the way from [https://2016.igem.org/Team:XMU-China - 2016 XMU-China iGEM team]. They required components from the biosensor part | + | Not long after, a more distant call reached us all the way from [https://2016.igem.org/Team:XMU-China - 2016 XMU-China iGEM team]. They required components from the biosensor part of the LMU team 2014 in order to detect and kill ''Staphylococcus aureus''. |
===Our dutch neighbours=== | ===Our dutch neighbours=== | ||
| − | + | Last but not least, [https://2016.igem.org/Team:Groningen the team from Groningen] got in touch with us regarding yet another Biobrick for ''Bacillus subtilis'' from LMU 2012. | |
| + | |||
| + | ===Double Kill=== | ||
| + | But we did not only ship BioBricks, we also requested some ourselves. For our Kill-Switch, we asked iGEM Freiburg (2010, BBa_K404113) and Slovenia (2012, BBa_K782063) for their parts. Since both parts inherited a pstI mutation site, we repaired this and enhanced the part with an additional poly-A sequence. | ||
| + | </div> | ||
| + | |||
| + | =HowTo= | ||
| + | <div class="white-box"> | ||
| + | In order to properly showcase our project, we decided to use the hightech 3D animation software [http://www.autodesk.de/products/maya/overview Maya by Autodesk]. As we consider this an awesome way to show people what you have been doing all summer long, here's an introduction video on HowTo do it. | ||
| + | |||
| + | <html> | ||
| + | <iframe width="812" height="457" src="https://www.youtube.com/embed/rS00leAqId8" frameborder="0" allowfullscreen></iframe> | ||
| + | </html> | ||
| + | |||
| + | We hope this helps you to get started and as we really want to see more of these animations, we also added our "cell" template file for you to download: | ||
| + | |||
| + | [[File:Muc16_CellTemplate.pdf|'''Cell Template Download''']] | ||
| + | </div> | ||
=References= | =References= | ||
| + | <div class="white-box"> | ||
<references /> | <references /> | ||
| + | </div> | ||
{{LMU-TUM_Munich_html_end}} | {{LMU-TUM_Munich_html_end}} | ||
Latest revision as of 12:19, 1 December 2016

Contents
Scientific Collaborations
Collaboration with Sheffield iGEM Team
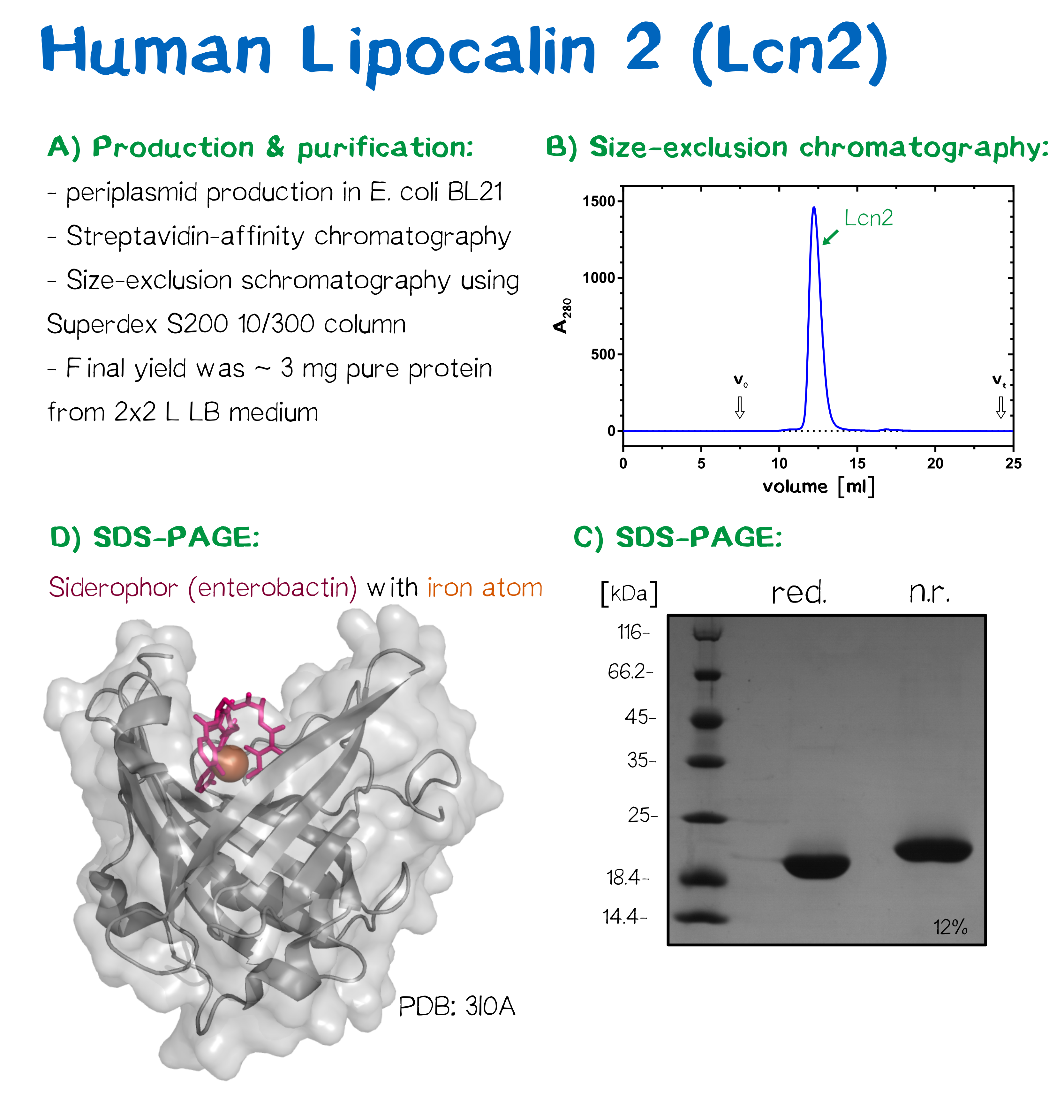
Collaboration with iGEM Sheffield : recombinant Lcn2
A) Production and purification strategy, B) Size-exclusion chromatography shows a monodisperse protein, C) the purified Lcn2 protein is >98% pure under reducing and non-reducing conditions in SDS-PAGE, D) crystal structure of Lcn2 (PDB: [http://www.rcsb.org/pdb/explore.do?structureId=3i0a| 3I0A]) together with the bound siderophore enterobactin and an iron atom.
We met the iGEM Team from Sheffield University at the European Experience in Paris where we presented our projects to each other. When they told us that they were using the human Lipocalin 2 (Lcn2) in order to detect the siderophores of pathogenic bacteria, we came up with the idea that we could provide the team with highly pure Lcn2 protein. This was a fruitful collaboration as our team's main PI, Prof. Dr. Arne Skerra, is one of the leading experts in the field of lipocalins[1]. For this reason, the expression constructs and the protocols to produce this recombinant protein are well established our affiliated lab. The main challenge in producing the recombinant protein was that the Sheffield iGEM team required the apo form of the protein without the bacterial siderophore as they aimed to scavenge siderophores from samples and then determine the siderophore content. Thus, we produced the recombinant protein in E. coli such that it was free from the siderophore generated in conventional E. coli strains.
Collaboration with Erlangen iGEM Team
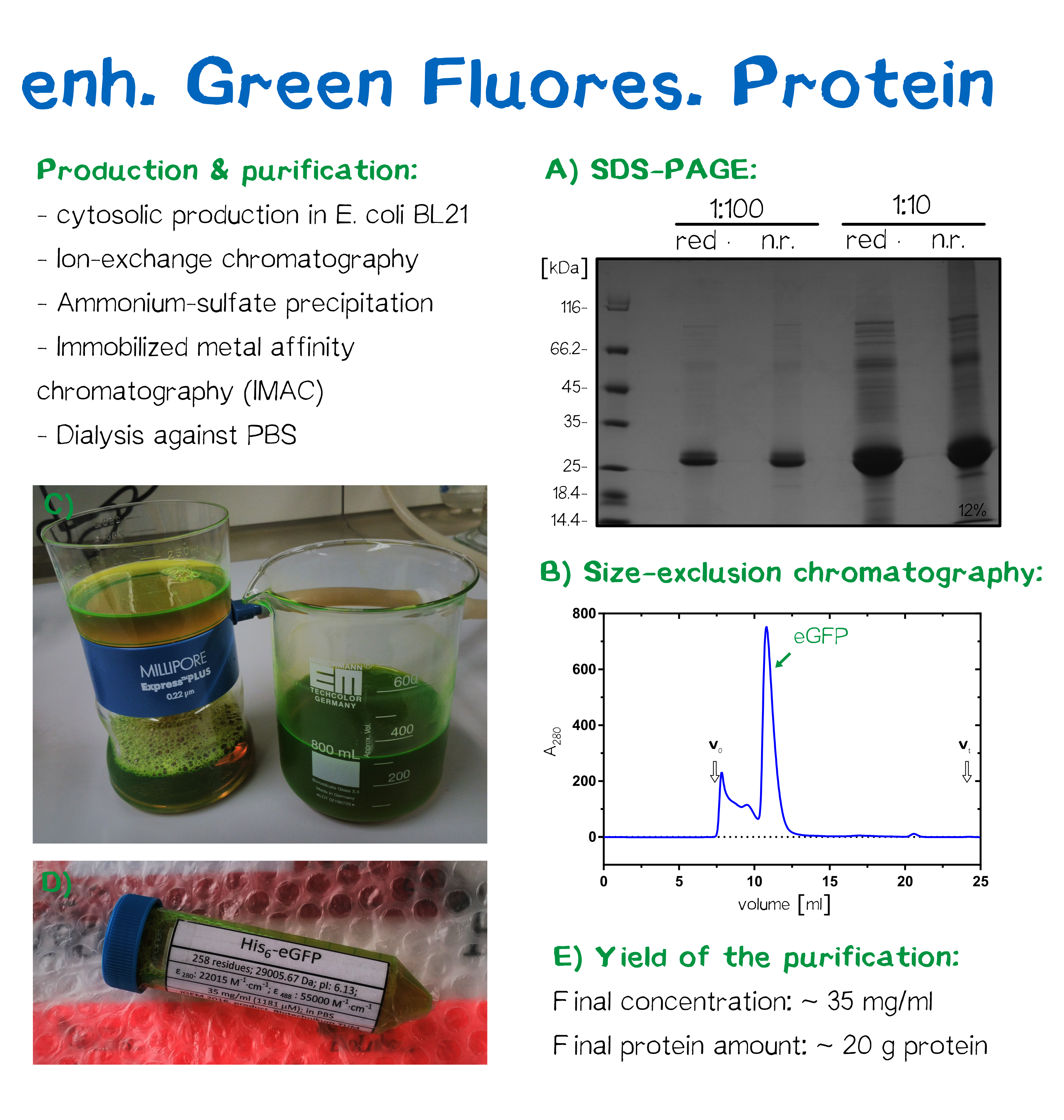
Collaboration with iGEM FAU Erlangen:
A) Production and purification strategy, B) Size-exclusion chromatography, C) Filtration, D) Tube readied for shipment, E) Yield after purification
The iGEM team from FAU Erlangen was working on the development of a biological photovoltaic cell, for which they aimed to use fluorescent proteins to harvest the photons. We saw their presentation at the iGEM Germany meet-up in Marburg, and as we used the purified recombinant eGFP for our own project in any case, we proposed to provide them with eGFP for their experiments. Luckily, at the "Biotechnikum" of the TUM, an annual lab course for bioprocess engineering students produces a large batch of eGFP in E. coli at a bioprocess scale of several hundred liters of culture medium. For our project, we were provided with ≈50 g of eGFP that we subsequently purified by ammonium sulfate precipitation as well as immobilized metal affinity chromatography (IMAC), and eventually dialyzed the protein against PBS. At the beginning of October, we provided 1.4 g of purified eGFP to the FAU Erlangen iGEM team.
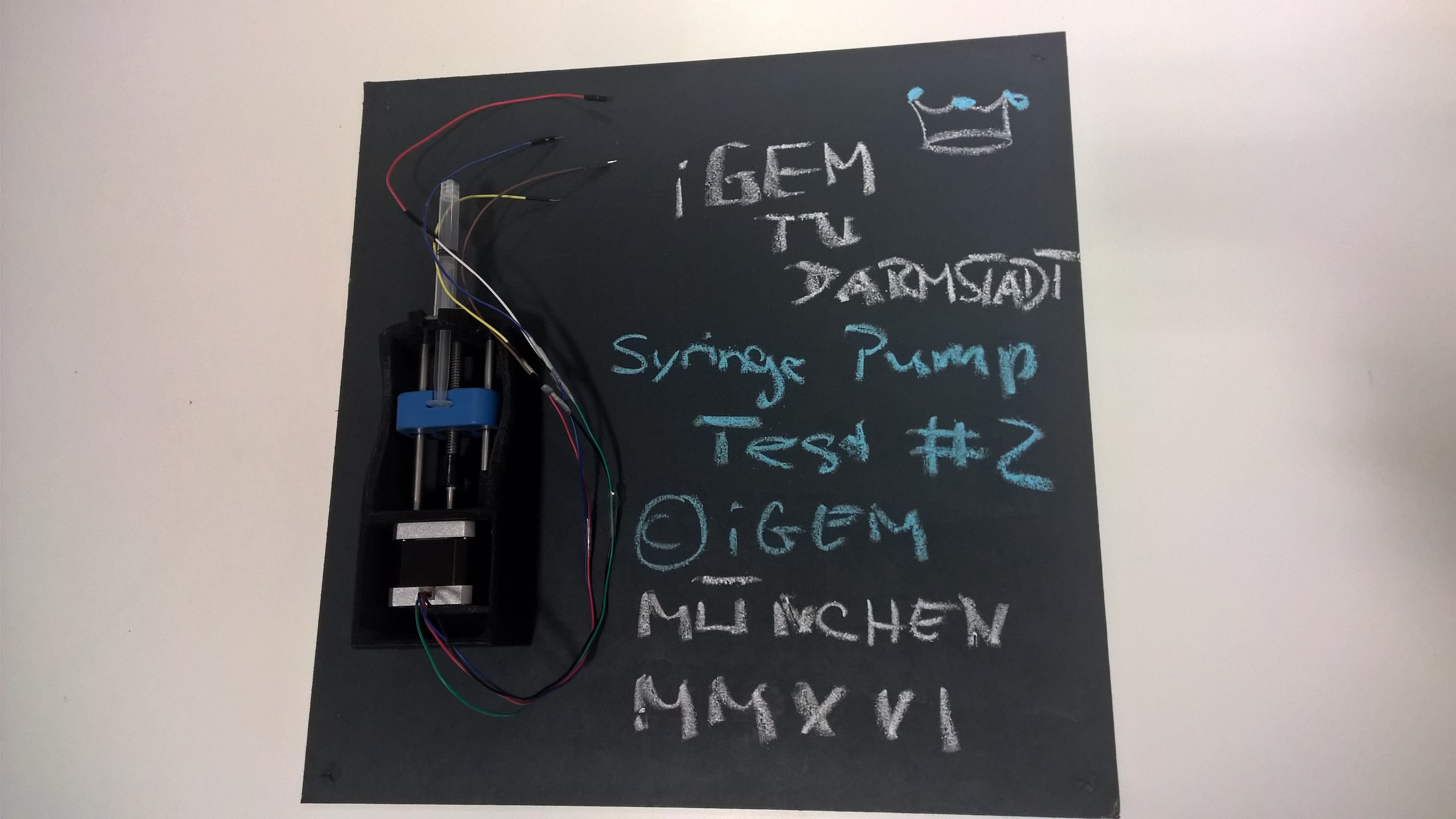
Collaboration with TU Darmstadt iGEM Team
In order to obtain first feedback for our DIY bioprinter kit, the iGEM Team from the TU Darmstadt agreed to become our beta-tester for the syringe pump that we had constructed for our bioprinter as well as for the whole Synbio community. Our syringe pump was proven to work well in our lab and it can be built with an investment of less than 50 USD provided an off-the-shelf 3D printer is available. For the beta test, the Darmstadt team received only the files for the 3D prints as well as all the parts (such as a stepper motor, few screws, some end-stop switches and the necessary wiring).
Here is their feedback from after they assembled our syringe pump:
"The pump is neat and stable. The idea of using buttons as end stops is, in principle, good. You took the necessary holes into account and we were able to assemble it according to the instructions. The click system for the syringe was well made and worked straightaway. The linear bearing could be used right away as well. We utilized a homemade 3D printer on highest gear for printing the parts. As you might know, this leads to relatively many "aberrations" and errors in the prints. They made the assembly difficult and we had to post-process several of them.
Your construction is designed quite closed, which renders it difficult for any passionate tinkerer to make adaptations after the printing. For instance, we noted that you had sent us a printed clutch though the stepper motor was not shortened and did not fit. Therefore, we drilled a hole in the front, enlarged the clutch and glued the parts together. In general, there are different stepper motors in use in the DIY community; hence, we suggest to conceptualize a more general clutch to ease this problem. Furthermore, the suspension device for the syringe could be improved, though the varying design of DIY printers makes finding a "good" solution rather difficult."
Meeting the iGEM community
The European Experience, Paris July 2nd to 3rd
The first big journey for our team lead us all the way to Paris, France. Following the invitation by the teams from iGEM IONIS and Evry, we packed our suitcases and were off.
And after a little climbing tour to Montmartre and maybe some super-last-minute poster printing ;), we settled for the night in our shady "lodging" to be ready to showcase our project publicly for the first time.
Early morning, we headed to the event location. First objective: COFFEE,
which ended up being a greater challenge than could have been anticipated...
Don't bring a knife (instant coffee) to a gun fight (horde of thirsty, tired students) :D
After breakfast was acquired, things started to get interesting. Dozens of awesome projects from all over Europe and even the UK! It was great to see all those ideas and of course to be able to showcase our own bioprinting project ourselves. We also obtained helpful tips and hints. After the poster sessions was over and nutrition had been taken care of, interesting panels on ethics and entrepreneurship in synthetic biology followed before it was time for Randy to drop some final Brexit jokes and conclude the scientific section of the event.
The evening promised to be a great dance party in "downtown Paris," including a live stream of the quarter-final match of the European Championship between Italy and of course Germany. After this sweat-breaking, super-tight, overly nerve-wracking game was finally over, with the better end for us :), we hit the dance floor down in the underground car park. The midnight surprise: a breathtaking fire-breather, who for some reason was interrupted by the security guard, we wonder why ;). With this awesome party night marking the end of the [http://www.facebook.com/events/458143747717965/ European Experience], we used the following Sunday to enjoy a wonderful French breakfast and explore the tourist sites of the "City of Love" before it was time to board the plane back to the lab. What a great event!
iGEM meets Marburg, Marburg August 5th to 7th
Just one month after the European Experience it was time for the next big meetup, the German meetup in the historic city Marburg with its winding streets and steep and tiny alleys.
Instead of boarding a plane, we loaded the car and went on a little road trip of 450 km through wind and rain. Luckily the weather got better and better the closer we came. Just after arriving at the camp site and setting everything up, the first official point of the program was already about to begin: bar tour, yeah! A long and noisy night followed ;) The next morning, the teams from literally all over Germany and extended Germany, also known as Denmark, assembled at the main event building and grabbed some awesome breakfast, including appropriate amounts of coffee, before being welcomed by our great hosts from [http://www.facebook.com/igemmarburg iGEM Marburg]. After introduction speeches by the sponsors of the event, the teams took the stage and presented their projects. As some time had passed since the Paris event, we got to see how the ideas had developed, and there were also some we had not seen before. Really awesome! After grabbing lunch, it was our turn to show what we had done and would still do in the lab. The afternoon was dedicated to physical activity of a special kind, Bubble Soccer. Probably the most hilarious game ever, though it will drain all your energy. Luckily our hosts had anticipated this and provided us with a tasty BBQ. Though everyone was a bit tired, the day was far from over as there was the final party of the weekend. The next morning came way too quickly, but luckily it was a beautiful sunny morning. We had the chance to hike to the castle and enjoy the amazing view from the top before it was again time to say Goodbye for a while and hit the road back to the lab. Thanks again Marburg for having us, it was a pleasure!
Helping other iGEM teams
iGEM experiment protocols in all languages by the METU HS Ankara iGEM team
This year's team of Highschool students from Ankara got in touch with us and asked for help for their clever idea of translating the standard iGEM lab protocols into different languages. This will hopefully help future teams from all around the globe to better understand them and apply them in the lab without being required to understand English perfectly. Especially other future Highschool teams should profit from this.
We are happy to be part such a great project and make iGEM even more international.
65 roses campaign by the iGEM team Tel Hai
As part of their project dealing with the most prevalent mutation in the CFTR protein, which is responsible for cystic fibrosis, the team from Tel Hai Academic college dedicated themselves to raise awareness for the severely life-shortening genetic condition. The “65 Roses” story itself dates back to 1965 when an observant 4-year-old, hearing the name of his disease for the first time, pronounced cystic fibrosis as "65 Roses." Nowadays, “65 Roses” is a term often used by young children with cystic fibrosis to pronounce the name of their disease. We are very happy to be a part of their noble efforts and dedication for this project.
BioBricks for the world
Due to the history of interesting projects from Munich, it did not take long for the first Biobricks requests to come in from all over the world.
Vive le France!
The first inquiry came from our cave-saving colleagues from Toulouse, who were in need of a [http://parts.igem.org/Part:BBa_K823026 BioBrick for their Bacillus subtilis] originally developed by the team from LMU in 2012. Go save those caves! ;)
The far east
Not long after, a more distant call reached us all the way from - 2016 XMU-China iGEM team. They required components from the biosensor part of the LMU team 2014 in order to detect and kill Staphylococcus aureus.
Our dutch neighbours
Last but not least, the team from Groningen got in touch with us regarding yet another Biobrick for Bacillus subtilis from LMU 2012.
Double Kill
But we did not only ship BioBricks, we also requested some ourselves. For our Kill-Switch, we asked iGEM Freiburg (2010, BBa_K404113) and Slovenia (2012, BBa_K782063) for their parts. Since both parts inherited a pstI mutation site, we repaired this and enhanced the part with an additional poly-A sequence.
HowTo
In order to properly showcase our project, we decided to use the hightech 3D animation software [http://www.autodesk.de/products/maya/overview Maya by Autodesk]. As we consider this an awesome way to show people what you have been doing all summer long, here's an introduction video on HowTo do it.
We hope this helps you to get started and as we really want to see more of these animations, we also added our "cell" template file for you to download:
References
- ↑ Schiefner, A., & Skerra, A. (2015). The menagerie of human lipocalins: a natural protein scaffold for molecular recognition of physiological compounds. Accounts of chemical research, 48(4), 976-985.