(→CMV promoter) |
VolkerMorath (Talk | contribs) |
||
| (85 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | {{LMU-TUM_Munich|navClass= | + | {{LMU-TUM_Munich|navClass=design}} |
| − | + | <div class="imagelink float-right">[[Media:Muc16_Sticker_Design_001.png]][[Image:Muc16_Sticker_Design_001.png|350px|link=]] | |
| − | [[ | + | </div> |
| − | + | ||
| − | In the following, we | + | =Receptor design highlights= |
| + | <div class="white-box"> | ||
| + | ''Think before you ink'' may be a popular slogan wielded by tattoo adversaries, but it has also been perfectly applicable to creating our synthetic biology approach of bioprinting. In the following, we explore the contemplations that contributed to the design of the membrane proteins (also referred to as 'receptors') that allow us to embed eukaryotic and bacterial cells into a [https://2016.igem.org/Team:LMU-TUM_Munich/Proof stable, three-dimensional and user-definable matrix] immediately upon printing. | ||
| + | </div> | ||
| − | + | =Human cell surface receptors= | |
| − | 1 | + | <div class="white-box"> |
| + | <div class="imagelink float-right">[[Media:Muc16 receptor schematic_002.png]][[Image:Muc16 receptor schematic_002.png|300px|link=]] | ||
| + | <div class="caption">'''Figure 1''': A) Receptor constructs in a schematic representation of functional genetic elements. B) Schematic representation of a receptor presenting an extracellular single-chain avidin that is endogenously biotinylated by a coexpressed biotin ligase (BirA) and thus presents biotin groups at the cell surface.</div></div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | For this purpose, we built '''two kinds of receptor constructs''', both on their own able – when expressed in mammalian cells – to cross-link cells for our approach of bioprinting: | |
| − | + | 1) '''A receptor presenting an extracellular biotin acceptor peptide''' that is endogenously biotinylated by a coexpressed biotin ligase (BirA), thus presenting biotin molecules on the cellular surface and allowing it to interact with streptavidin in the printing solution. | |
| − | + | 2) '''Receptors presenting an extracellular monomeric or single-chain avidin variant'''. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | In presenting avidin on the cell surface, cells are able to interact with a co-injected biotinylated linker peptide. With this linker connected to several cell surface avidins as well as streptavidin in the printing solution, a protein matrix is created that embeds cells and allows precise positioning of cells in a three-dimensional manner. | |
| − | + | Our chosen avidin derivates by design allow functional fusion of the otherwise tetrameric avidin molecule with our receptor. Two different variants were used: ''enhanced monomeric avidin'', a single-subunit avidin that is nevertheless able to bind biotin, and ''single-chain avidin'', which resembles the naturally occurring avidin tetramer but consisting of a single polypeptide chain. | |
| − | + | All receptors mediate the same function: By interacting with streptavidin in the printing reservoir – either directly by presenting biotin (biotin acceptor peptide) or indirectly via binding to a biotinylated linker (single-chain avidin variants), cells are cross-linked due to polyvalent binding of streptavidin molecules in the printing solution to several cellular biotin groups at once. | |
| − | + | ||
</div> | </div> | ||
| − | == | + | ==The incredible journey: signal peptides and protein targeting== |
| − | + | ||
<div class="white-box"> | <div class="white-box"> | ||
| − | + | Essential to a protein's function are not only its activity or binding properties, but also its presence at the right time in the right place – the latter in our case being the cell surface. One ubiquitous element for protein targeting is the signal peptide, which enables translocation of proteins to the ER, from where their journey continues to their place of function. Proteins containing a signal peptide include transmembrane proteins, secreted proteins, proteins of the ER itself, proteins of the Golgi apparatus and several more.<ref>Rapoport, T. A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature, 450(7170), 663-669.</ref> For our project, signal peptides constituted a crucial part of many constructs, used to target transmembrane proteins to the cell surface (receptors) and proteins to the ER (BirA), and to secrete proteins into the surrounding medium (luciferase assays to quantify expression levels). | |
| − | </ | + | |
| − | === | + | <div class="imagelink float-right">[[Media:Muc16 SP schematic.jpeg]][[Image:Muc16 SP schematic.jpeg|420px|link=]] |
| + | <div class="caption">'''Figure 2:''' Cotranslational translocation of a membrane protein into the ER in a schematic depiction showing the nascent, unfolded peptide chain containing signal peptide (''start-transfer sequence'') and ''stop-transfer sequence'' as well as the ER membrane.<ref>Taken from Alberts, B., Bray, D., Hopkin, K., Johnson, A., Lewis, J., Raff, M., ... & Walter, P. (2010). Essential cell biology. Garland Science.</ref></div></div> | ||
| − | + | The hydrophobic signal peptide (also called ''start-transfer sequence'') usually consists of less than 30 amino acids <ref>Walter, P., Ibrahimi, I., & Blobel, G. U. N. T. E. R. (1981). Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. The Journal of Cell Biology, 91(2), 545-550.</ref> and is recognized by the signal recognition particle upon translation at the rough ER, mediating cotranslational translocation of the nascent peptide chain into the ER.<ref>Keenan, Robert J., et al. "The signal recognition particle." Annual review of biochemistry 70.1 (2001): 755-775.</ref> | |
| − | The | + | |
| − | + | During this project, three different signal peptide constructs were tested in order to determine that resulting in the highest expression levels: | |
| − | ''' | + | * The '''EGFR signal peptide''' was taken from the iGEM parts registry ([http://parts.igem.org/Part:BBa_K157001:Design BBa_K157001]) and combined with a BioBrick containing the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard. |
| − | + | * The '''BM40 and Igκ signal peptides''' were designed by us and synthesized with regard to adding additional spacing and functional elements to the 5'-UTR of the constructs, and combined with a BioBrick containing the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard. | |
| − | + | ||
| − | + | ||
| + | The EGFR signal peptide taken from the parts registry is constructed such that the signal peptide ORF immediately follows the RFC10 cloning scar after the CMV promoter, thus resulting in a very short 5’ untranslated region (UTR) of transcribed mRNA. The combination of the CMV promoter with the synthesized BM40 and Igκ signal peptides allows for a considerably longer 5’-UTR – and can thus be made to contain a full Kozak consensus sequence by design. The Kozak sequence is recognized by the ribosome as a translational start site; if this element is missing or deviates from the consensus sequence, this may considerably decrease translation efficiency.<ref>Kozak, M. (1989). The scanning model for translation: an update. The Journal of cell biology, 108(2), 229-241.</ref> For the EGFR signal peptide construct, a Kozak sequence is not present, as it would have to precede the start codon ATG, i.e. the position occupied by the RFC10 cloning scar. | ||
| + | Since both the distance from the promoter to the open reading frame and the Kozak consensus sequence are considered crucial parameters for expression levels<ref>Kozak, M. (1987). An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic acids research, 15(20), 8125-8148.</ref>, BM40 and Igκ signal peptide constructs are likely to result in increased expression levels compared to proteins containing the EGFR signal peptide from the parts registry. | ||
| + | </div> | ||
<div class="white-box"> | <div class="white-box"> | ||
| − | + | In order to predict whether our designed signal peptides would be functional, we used the [http://www.cbs.dtu.dk/services/SignalP SignalP] server, which allows to make theoretical predictions about signal peptide functionality and cleavage.<ref>Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods, 8(10), 785-786.</ref> | |
| + | |||
| + | <div class="imagelink float-right">[[Media:Muc16 SignalPeptideprediction_002.png]][[Image:Muc16 SignalPeptideprediction_002.png|820px|link=]] | ||
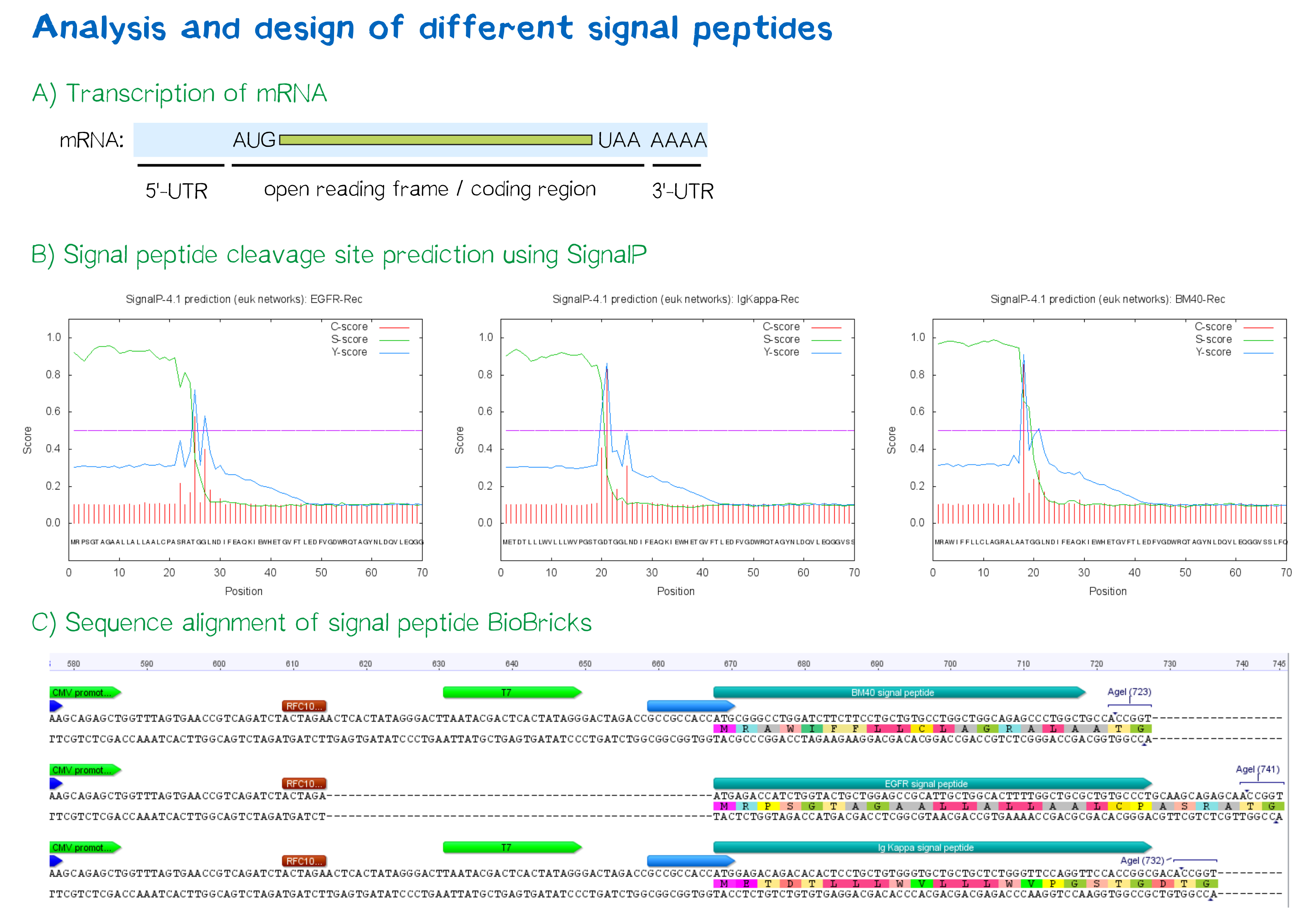
| + | <div class="caption">'''Figure 3:''' A) Schematic depiction of a protein-coding mRNA containing a 5'- and 3'-UTR, the first of which is an important factor in translation efficiency. B) Theoretical prediction of signal peptides within the receptor construct containing the EGFR, Igκ and BM40 signal peptide via the SignalP 4.1 server. Plotted probabilities represent the raw cleavage site “C-score”, the signal peptide “S-score” and the combined cleavage site “Y-score”. C) ClustalW multiple sequence alignment of the BM40, EGFR and Igκ signal peptide construct, depicting a gap for the EGFR signal peptide construct compared to the newly designed ones and thus indicating a shorter length of the 5'-UTR . For Igκ and BM40 signal peptide constructs, a T7 promoter spacer as well as a Kozak sequence increase the distance between TATA box and protein-coding region.</div></div> | ||
| − | |||
</div> | </div> | ||
| − | == | + | ==Elements for stability and detection== |
<div class="white-box"> | <div class="white-box"> | ||
| Line 71: | Line 65: | ||
<div class="white-box"> | <div class="white-box"> | ||
| − | For anchoring in the membrane, | + | For anchoring in the membrane, a type I membrane protein with defined localization of N- and C-terminus out- and inside of the cell was used. The N-terminal transmembrane α-helix of human EGFR (UniProt P00533, amino acids 622-653) was herefore chosen (see below the topology prediction of EGFR via the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server for the prediction of transmembrane helices]<ref>Sonnhammer, E. L., Von Heijne, G., & Krogh, A. (1998, July). A hidden Markov model for predicting transmembrane helices in protein sequences. In Ismb (Vol. 6, pp. 175-182).</ref>). A stop-transfer sequence consisting of charged amino acids - being characteristic for type I membrane proteins - was added at the C-terminus, and the sequence was furthermore flanked by a (GGGGC)<sub>2</sub>-linker at the N- and C-terminus, respectively. The addition of a stop-transfer sequence as in the naturally occurring EGFR sequence, as well as the addition of flexible linkers, makes our EGFR-TMD an improvement over the BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002], which does not possess either of these elements. |
| − | [[ | + | <div class="imagelink float-right">[[Media:Receptor TMD 002.png]][[Image:Receptor TMD 002.png|820px|link=]] |
| + | <div class="caption"></div>'''Figure 4:''' A) Multiple sequence alignment of human EGFR (see [http://www.uniprot.org/uniprot/P00533 UniProt P00533]), the EGFR transmembrane domain BioBrick from the registry ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002] and the extended EGFR transmembrane designed by us, containing a charged stop-transfer domain as well as linker elements at the C- and N-termini. B) Transmembrane domain prediction by the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server], indicating positioning of N- and C-termini out- and inside of the cell, respectively, as well as predicting functionality of transmembrane regions. C) Schematic depiction of the C-terminal stop-transfer sequence of the receptor.</div> | ||
| + | </div> | ||
| + | ==Elements for detection== | ||
| + | <div class="white-box"> | ||
| + | In order to confirm the correct expression and localization of the receptor, the receptor contains '''three functional elements for its detection''' for both the N- and the C-terminal receptor domains: | ||
| − | + | * The intracellularly located C-terminal '''red fluorescent protein mRuby 3''' for detection of the receptor via fluorescence microscopy while also providing a stable, folded domain at the C-terminus, | |
| + | * an '''extracellular epitope domain''' near the N-terminus for immunochemical detection via A3C5-antibody fragments as well as FACS, and | ||
| + | * an '''intracellular <i>Strep</i>-tag II''' at the N-terminus for the detection and purification via immunochemical methods. | ||
| − | + | Further elements that were included into the expression construct are the CMV promoter for overall high expression levels of the receptor, resulting in a high-avidity-interaction. At the 3'-end, the construct contains the polyadenylation signal of human growth hormone (hGH) for functional polyadenylation of the transcribed mRNA. | |
</div> | </div> | ||
| − | = | + | =Design of an autotransporter construct for bacterial surface display= |
<div class="white-box"> | <div class="white-box"> | ||
| − | + | Besides the design of biotinylated and biotin-binding receptors for eukaryotic cells, we also wanted to apply our <span style="color:#009440">biot</span><span style="color:#3070b3">INK</span> approach for prokaryotic cells. For this purpose, we designed an autotransporter device that is able to present biotinylated or biotin-binding protein domains on the surface of ''E. coli''. Constructs for bacterial surface display are hereby already well known in the field of protein engineering of therapeutic proteins, such as antibodies. There, they are used to screen libraries of different antibodies concerning their affinity towards a given therapeutic target. | |
| − | + | ||
| − | + | We based our design on the autotransporter EspP<ref>Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.</ref>. | |
| − | + | <div class="imagelink float-right">[[Media:Muc16 Autotransporterdesign_001.png]][[Image:Muc16 Autotransporterdesign_001.png|820px|link=]] | |
| − | + | <div class="caption"></div>'''Figure 5'''</div> | |
| − | </div> | + | |
| − | + | The autotransporter system itself is established and functional (see Fig. A<ref>Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.</ref>), and here we "bricked" it down into RFC[10] and RFC[25] BioBricks. At first, we tested different inducible bacterial promoter systems (e.g. "TetR repressed GFP"; [http://parts.igem.org/Part:BBa_K577893 BBa_K577893]) that are available in the parts registry, but when we saw that two of the available systems did not work in our own hands, we decided to built the system on our own. We used the Tet-Repressor generator that was available on the distribution plates ([http://parts.igem.org/Part:BBa_I739001 BBa_I739001]) as we believe that it is very important for the standardization in SynBio to use available parts. Downstream of this repressor generator we designed and fused a BioBrick ([http://parts.igem.org/Part:BBa_K2170141 BBa_K2170141]) that encodes a double Tet-Operator (Tet-Repressor binding site together with a bacterial promoter and an signal peptide of the outer membrane protein A (OmpA<ref>Ghrayeb, J., Kimura, H., Takahara, M., Hsiung, H., Masui, Y., & Inouye, M. (1984). Secretion cloning vectors in Escherichia coli. The EMBO journal, 3(10), 2437.</ref>) from ''E. coli'' that has a 3' RFC[25] restriction site, allowing the protein fusion of proteins that are secreted into the bacterial periplasm when expressed with this BioBrick. As a protein fusion, we then assembled a protein cargo domain that is generally exchangeable for any protein that is so small in size that it can be transported through the outer membrane by the EspP autotransporter. In our project we used the same extracellular domains as for the eukaryotic receptors: enhanced monomeric Avidin (eMA; [http://parts.igem.org/Part:BBa_K2170204 BBa_K2170204]), single-chain Avidin (scAvidin; [http://parts.igem.org/Part:BBa_K2170205 BBa_K2170205]) and a composite part composed of a N-terminal Biotinylation acceptor peptide (BAP) and a C-terminal NanoLuciferase (together shown as BAP; ; [http://parts.igem.org/Part:BBa_K2170118 BBa_K2170118], see Fig. B). Our design features an A3C5-epitope tag further downstream that can be recognized by a high-affinity Fab-fragment and is generally used to detect and quantify the surface display of the cargo domain<ref>Costa, J., Grabenhorst, E., Nimtz, M., & Conradt, H. S. (1997). Stable expression of the Golgi form and secretory variants of human fucosyltransferase III from BHK-21 cells Purification and characterization of an engineered truncated form from the culture medium. Journal of Biological Chemistry, 272(17), 11613-11621.</ref>. Further downstream of this affinity tag we fused the BioBrick for the EspP autotransporter from ''E. coli''<ref>Barnard, T. J., Dautin, N., Lukacik, P., Bernstein, H. D., & Buchanan, S. K. (2007). Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nature structural & molecular biology, 14(12), 1214-1220.</ref><ref>Skillman, K. M., Barnard, T. J., Peterson, J. H., Ghirlando, R., & Bernstein, H. D. (2005). Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Molecular microbiology, 58(4), 945-958.</ref> which is then followed by a bacterial terminator. For the termination we chose again a well-working terminator from the distribution plate ("double terminator"; [http://parts.igem.org/Part:BBa_B0010 BBa_B0010]-[http://parts.igem.org/Part:BBa_B0012 BBa_B0012]).<br> But this was just the DNA-part. What is expected to happen on a protein level? The construct constantly produces Tet-repressor that binds to the Tet-Operator and repressed the promoter activity of the autotransporter gene. As soon as the inducer anhydrotetracycline (aTc) is added to the culture, it binds to the Tet-repressor that can't bind anymore to the Tet-operator and thus the expression of the autotransporter can be regulated. Although the TetR-system is known to be a tight promoter system there is always a certain background expression with most promoters. If the protein expression of the autotransporter is induced using aTc the autotransporter is transcribed and translated. Due to the bacterial signal peptide, the protein is secreted into the bacterial periplasm (the space between the two bacterial membranes of gram-negative bacteria such as ''E. coli''). When present in the bacteria periplasm the autotransporter diffused to the outer membrane and inserts into the membrane. After the integration into the outer membrane, the protein cargo is transported through the beta-barrel of the autotransporter and is presented on the bacterial surface. We are sure that this BioBrick will be a valuable contribution to the Parts Registry as it allows future teams to display small protein domains on the surface of gram-negative bacteria, such as ''E. coli''. | |
| − | + | ||
| − | + | ||
</div> | </div> | ||
| − | = | + | =References= |
<div class="white-box"> | <div class="white-box"> | ||
| − | + | <references /> | |
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{LMU-TUM_Munich_html_end}} | {{LMU-TUM_Munich_html_end}} | ||
Latest revision as of 14:10, 3 December 2016
Contents
Receptor design highlights
Think before you ink may be a popular slogan wielded by tattoo adversaries, but it has also been perfectly applicable to creating our synthetic biology approach of bioprinting. In the following, we explore the contemplations that contributed to the design of the membrane proteins (also referred to as 'receptors') that allow us to embed eukaryotic and bacterial cells into a stable, three-dimensional and user-definable matrix immediately upon printing.
Human cell surface receptors
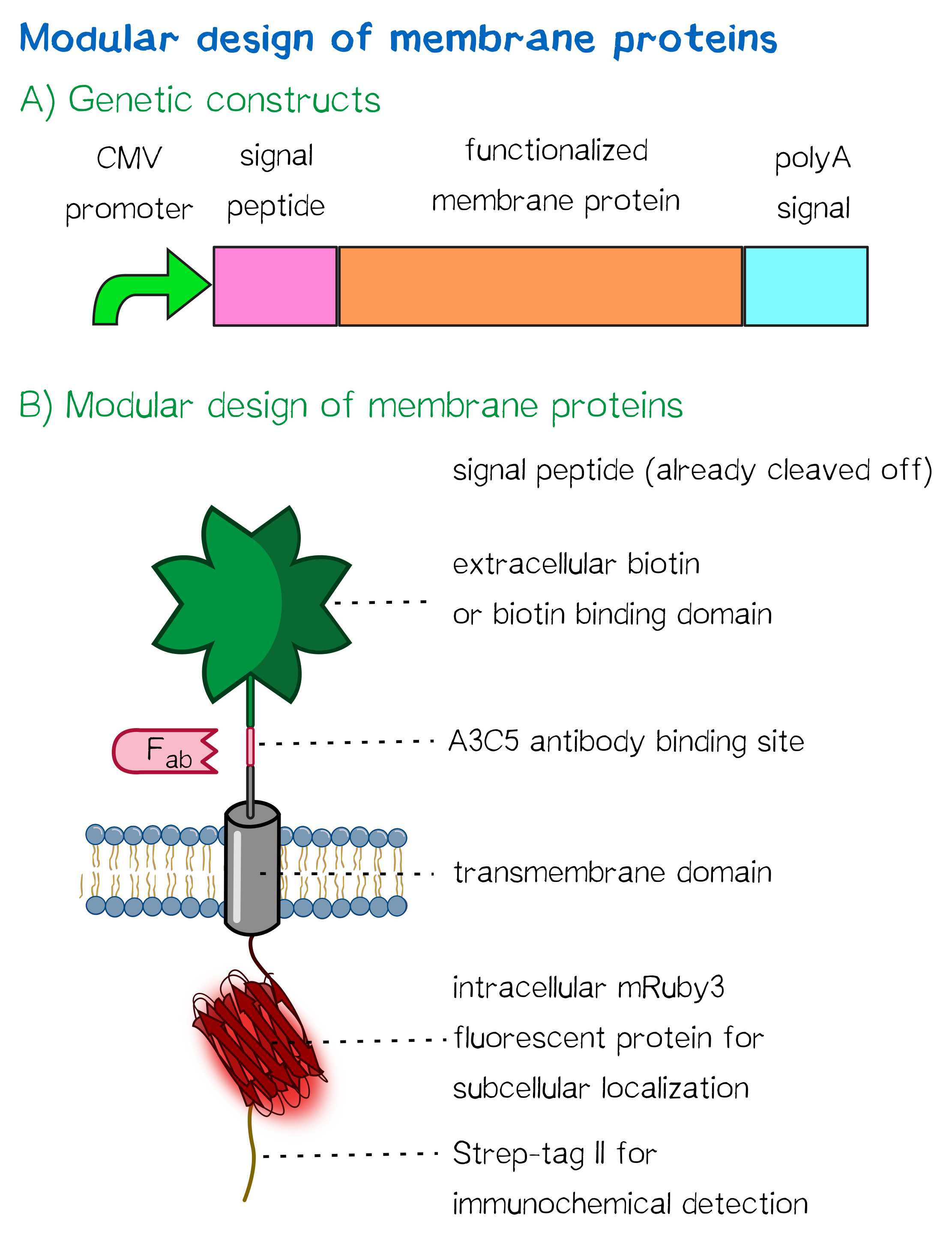
For this purpose, we built two kinds of receptor constructs, both on their own able – when expressed in mammalian cells – to cross-link cells for our approach of bioprinting:
1) A receptor presenting an extracellular biotin acceptor peptide that is endogenously biotinylated by a coexpressed biotin ligase (BirA), thus presenting biotin molecules on the cellular surface and allowing it to interact with streptavidin in the printing solution.
2) Receptors presenting an extracellular monomeric or single-chain avidin variant.
In presenting avidin on the cell surface, cells are able to interact with a co-injected biotinylated linker peptide. With this linker connected to several cell surface avidins as well as streptavidin in the printing solution, a protein matrix is created that embeds cells and allows precise positioning of cells in a three-dimensional manner.
Our chosen avidin derivates by design allow functional fusion of the otherwise tetrameric avidin molecule with our receptor. Two different variants were used: enhanced monomeric avidin, a single-subunit avidin that is nevertheless able to bind biotin, and single-chain avidin, which resembles the naturally occurring avidin tetramer but consisting of a single polypeptide chain.
All receptors mediate the same function: By interacting with streptavidin in the printing reservoir – either directly by presenting biotin (biotin acceptor peptide) or indirectly via binding to a biotinylated linker (single-chain avidin variants), cells are cross-linked due to polyvalent binding of streptavidin molecules in the printing solution to several cellular biotin groups at once.
The incredible journey: signal peptides and protein targeting
Essential to a protein's function are not only its activity or binding properties, but also its presence at the right time in the right place – the latter in our case being the cell surface. One ubiquitous element for protein targeting is the signal peptide, which enables translocation of proteins to the ER, from where their journey continues to their place of function. Proteins containing a signal peptide include transmembrane proteins, secreted proteins, proteins of the ER itself, proteins of the Golgi apparatus and several more.[1] For our project, signal peptides constituted a crucial part of many constructs, used to target transmembrane proteins to the cell surface (receptors) and proteins to the ER (BirA), and to secrete proteins into the surrounding medium (luciferase assays to quantify expression levels).
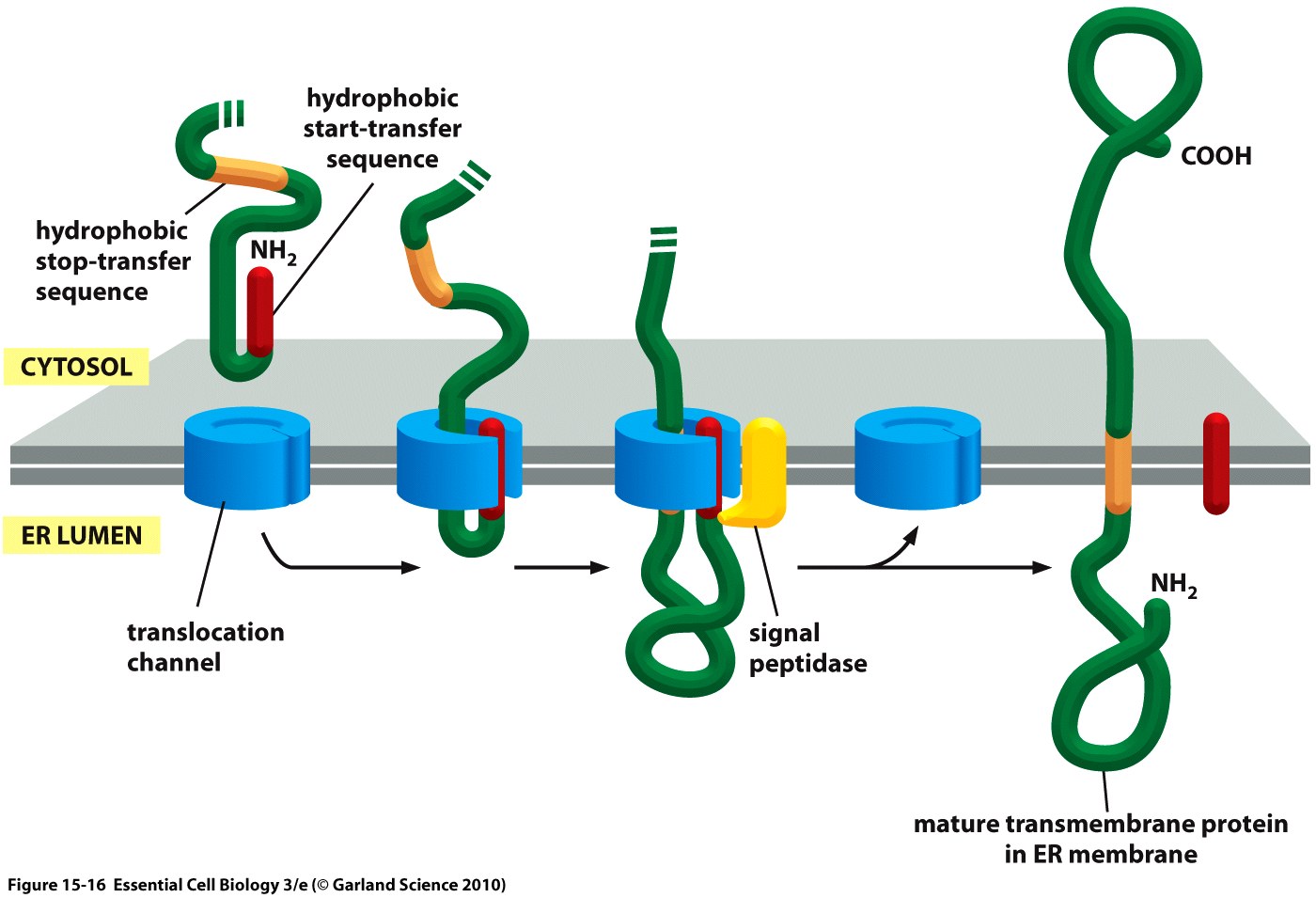
The hydrophobic signal peptide (also called start-transfer sequence) usually consists of less than 30 amino acids [3] and is recognized by the signal recognition particle upon translation at the rough ER, mediating cotranslational translocation of the nascent peptide chain into the ER.[4]
During this project, three different signal peptide constructs were tested in order to determine that resulting in the highest expression levels:
- The EGFR signal peptide was taken from the iGEM parts registry ([http://parts.igem.org/Part:BBa_K157001:Design BBa_K157001]) and combined with a BioBrick containing the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard.
- The BM40 and Igκ signal peptides were designed by us and synthesized with regard to adding additional spacing and functional elements to the 5'-UTR of the constructs, and combined with a BioBrick containing the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard.
The EGFR signal peptide taken from the parts registry is constructed such that the signal peptide ORF immediately follows the RFC10 cloning scar after the CMV promoter, thus resulting in a very short 5’ untranslated region (UTR) of transcribed mRNA. The combination of the CMV promoter with the synthesized BM40 and Igκ signal peptides allows for a considerably longer 5’-UTR – and can thus be made to contain a full Kozak consensus sequence by design. The Kozak sequence is recognized by the ribosome as a translational start site; if this element is missing or deviates from the consensus sequence, this may considerably decrease translation efficiency.[5] For the EGFR signal peptide construct, a Kozak sequence is not present, as it would have to precede the start codon ATG, i.e. the position occupied by the RFC10 cloning scar.
Since both the distance from the promoter to the open reading frame and the Kozak consensus sequence are considered crucial parameters for expression levels[6], BM40 and Igκ signal peptide constructs are likely to result in increased expression levels compared to proteins containing the EGFR signal peptide from the parts registry.
In order to predict whether our designed signal peptides would be functional, we used the [http://www.cbs.dtu.dk/services/SignalP SignalP] server, which allows to make theoretical predictions about signal peptide functionality and cleavage.[7]
Elements for stability and detection
Apart from the functional parts that mediate the (strept)avidin-biotin interaction, other elements in the receptor were designed to make sure it is optimally translocated to the membrane, as stable as possible, and easily detectable.
EGFR transmembrane domain
For anchoring in the membrane, a type I membrane protein with defined localization of N- and C-terminus out- and inside of the cell was used. The N-terminal transmembrane α-helix of human EGFR (UniProt P00533, amino acids 622-653) was herefore chosen (see below the topology prediction of EGFR via the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server for the prediction of transmembrane helices][8]). A stop-transfer sequence consisting of charged amino acids - being characteristic for type I membrane proteins - was added at the C-terminus, and the sequence was furthermore flanked by a (GGGGC)2-linker at the N- and C-terminus, respectively. The addition of a stop-transfer sequence as in the naturally occurring EGFR sequence, as well as the addition of flexible linkers, makes our EGFR-TMD an improvement over the BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002], which does not possess either of these elements.
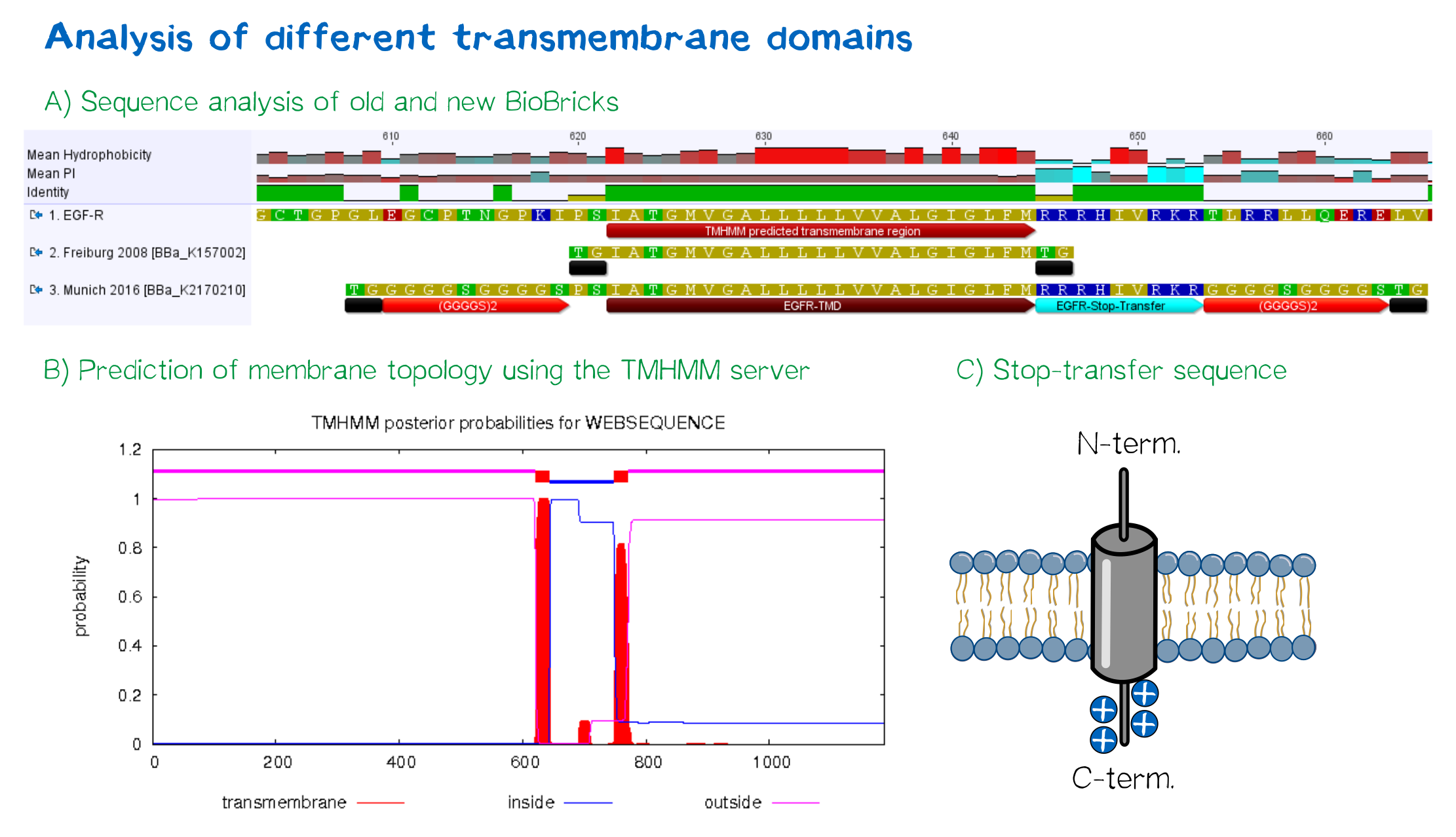 Figure 4: A) Multiple sequence alignment of human EGFR (see [http://www.uniprot.org/uniprot/P00533 UniProt P00533]), the EGFR transmembrane domain BioBrick from the registry ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002] and the extended EGFR transmembrane designed by us, containing a charged stop-transfer domain as well as linker elements at the C- and N-termini. B) Transmembrane domain prediction by the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server], indicating positioning of N- and C-termini out- and inside of the cell, respectively, as well as predicting functionality of transmembrane regions. C) Schematic depiction of the C-terminal stop-transfer sequence of the receptor.
Figure 4: A) Multiple sequence alignment of human EGFR (see [http://www.uniprot.org/uniprot/P00533 UniProt P00533]), the EGFR transmembrane domain BioBrick from the registry ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002] and the extended EGFR transmembrane designed by us, containing a charged stop-transfer domain as well as linker elements at the C- and N-termini. B) Transmembrane domain prediction by the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server], indicating positioning of N- and C-termini out- and inside of the cell, respectively, as well as predicting functionality of transmembrane regions. C) Schematic depiction of the C-terminal stop-transfer sequence of the receptor.Elements for detection
In order to confirm the correct expression and localization of the receptor, the receptor contains three functional elements for its detection for both the N- and the C-terminal receptor domains:
- The intracellularly located C-terminal red fluorescent protein mRuby 3 for detection of the receptor via fluorescence microscopy while also providing a stable, folded domain at the C-terminus,
- an extracellular epitope domain near the N-terminus for immunochemical detection via A3C5-antibody fragments as well as FACS, and
- an intracellular Strep-tag II at the N-terminus for the detection and purification via immunochemical methods.
Further elements that were included into the expression construct are the CMV promoter for overall high expression levels of the receptor, resulting in a high-avidity-interaction. At the 3'-end, the construct contains the polyadenylation signal of human growth hormone (hGH) for functional polyadenylation of the transcribed mRNA.
Design of an autotransporter construct for bacterial surface display
Besides the design of biotinylated and biotin-binding receptors for eukaryotic cells, we also wanted to apply our biotINK approach for prokaryotic cells. For this purpose, we designed an autotransporter device that is able to present biotinylated or biotin-binding protein domains on the surface of E. coli. Constructs for bacterial surface display are hereby already well known in the field of protein engineering of therapeutic proteins, such as antibodies. There, they are used to screen libraries of different antibodies concerning their affinity towards a given therapeutic target.
We based our design on the autotransporter EspP[9].
The autotransporter system itself is established and functional (see Fig. A[10]), and here we "bricked" it down into RFC[10] and RFC[25] BioBricks. At first, we tested different inducible bacterial promoter systems (e.g. "TetR repressed GFP"; [http://parts.igem.org/Part:BBa_K577893 BBa_K577893]) that are available in the parts registry, but when we saw that two of the available systems did not work in our own hands, we decided to built the system on our own. We used the Tet-Repressor generator that was available on the distribution plates ([http://parts.igem.org/Part:BBa_I739001 BBa_I739001]) as we believe that it is very important for the standardization in SynBio to use available parts. Downstream of this repressor generator we designed and fused a BioBrick ([http://parts.igem.org/Part:BBa_K2170141 BBa_K2170141]) that encodes a double Tet-Operator (Tet-Repressor binding site together with a bacterial promoter and an signal peptide of the outer membrane protein A (OmpA[11]) from E. coli that has a 3' RFC[25] restriction site, allowing the protein fusion of proteins that are secreted into the bacterial periplasm when expressed with this BioBrick. As a protein fusion, we then assembled a protein cargo domain that is generally exchangeable for any protein that is so small in size that it can be transported through the outer membrane by the EspP autotransporter. In our project we used the same extracellular domains as for the eukaryotic receptors: enhanced monomeric Avidin (eMA; [http://parts.igem.org/Part:BBa_K2170204 BBa_K2170204]), single-chain Avidin (scAvidin; [http://parts.igem.org/Part:BBa_K2170205 BBa_K2170205]) and a composite part composed of a N-terminal Biotinylation acceptor peptide (BAP) and a C-terminal NanoLuciferase (together shown as BAP; ; [http://parts.igem.org/Part:BBa_K2170118 BBa_K2170118], see Fig. B). Our design features an A3C5-epitope tag further downstream that can be recognized by a high-affinity Fab-fragment and is generally used to detect and quantify the surface display of the cargo domain[12]. Further downstream of this affinity tag we fused the BioBrick for the EspP autotransporter from E. coli[13][14] which is then followed by a bacterial terminator. For the termination we chose again a well-working terminator from the distribution plate ("double terminator"; [http://parts.igem.org/Part:BBa_B0010 BBa_B0010]-[http://parts.igem.org/Part:BBa_B0012 BBa_B0012]).
But this was just the DNA-part. What is expected to happen on a protein level? The construct constantly produces Tet-repressor that binds to the Tet-Operator and repressed the promoter activity of the autotransporter gene. As soon as the inducer anhydrotetracycline (aTc) is added to the culture, it binds to the Tet-repressor that can't bind anymore to the Tet-operator and thus the expression of the autotransporter can be regulated. Although the TetR-system is known to be a tight promoter system there is always a certain background expression with most promoters. If the protein expression of the autotransporter is induced using aTc the autotransporter is transcribed and translated. Due to the bacterial signal peptide, the protein is secreted into the bacterial periplasm (the space between the two bacterial membranes of gram-negative bacteria such as E. coli). When present in the bacteria periplasm the autotransporter diffused to the outer membrane and inserts into the membrane. After the integration into the outer membrane, the protein cargo is transported through the beta-barrel of the autotransporter and is presented on the bacterial surface. We are sure that this BioBrick will be a valuable contribution to the Parts Registry as it allows future teams to display small protein domains on the surface of gram-negative bacteria, such as E. coli.
References
- ↑ Rapoport, T. A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature, 450(7170), 663-669.
- ↑ Taken from Alberts, B., Bray, D., Hopkin, K., Johnson, A., Lewis, J., Raff, M., ... & Walter, P. (2010). Essential cell biology. Garland Science.
- ↑ Walter, P., Ibrahimi, I., & Blobel, G. U. N. T. E. R. (1981). Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. The Journal of Cell Biology, 91(2), 545-550.
- ↑ Keenan, Robert J., et al. "The signal recognition particle." Annual review of biochemistry 70.1 (2001): 755-775.
- ↑ Kozak, M. (1989). The scanning model for translation: an update. The Journal of cell biology, 108(2), 229-241.
- ↑ Kozak, M. (1987). An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic acids research, 15(20), 8125-8148.
- ↑ Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods, 8(10), 785-786.
- ↑ Sonnhammer, E. L., Von Heijne, G., & Krogh, A. (1998, July). A hidden Markov model for predicting transmembrane helices in protein sequences. In Ismb (Vol. 6, pp. 175-182).
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Ghrayeb, J., Kimura, H., Takahara, M., Hsiung, H., Masui, Y., & Inouye, M. (1984). Secretion cloning vectors in Escherichia coli. The EMBO journal, 3(10), 2437.
- ↑ Costa, J., Grabenhorst, E., Nimtz, M., & Conradt, H. S. (1997). Stable expression of the Golgi form and secretory variants of human fucosyltransferase III from BHK-21 cells Purification and characterization of an engineered truncated form from the culture medium. Journal of Biological Chemistry, 272(17), 11613-11621.
- ↑ Barnard, T. J., Dautin, N., Lukacik, P., Bernstein, H. D., & Buchanan, S. K. (2007). Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nature structural & molecular biology, 14(12), 1214-1220.
- ↑ Skillman, K. M., Barnard, T. J., Peterson, J. H., Ghirlando, R., & Bernstein, H. D. (2005). Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Molecular microbiology, 58(4), 945-958.


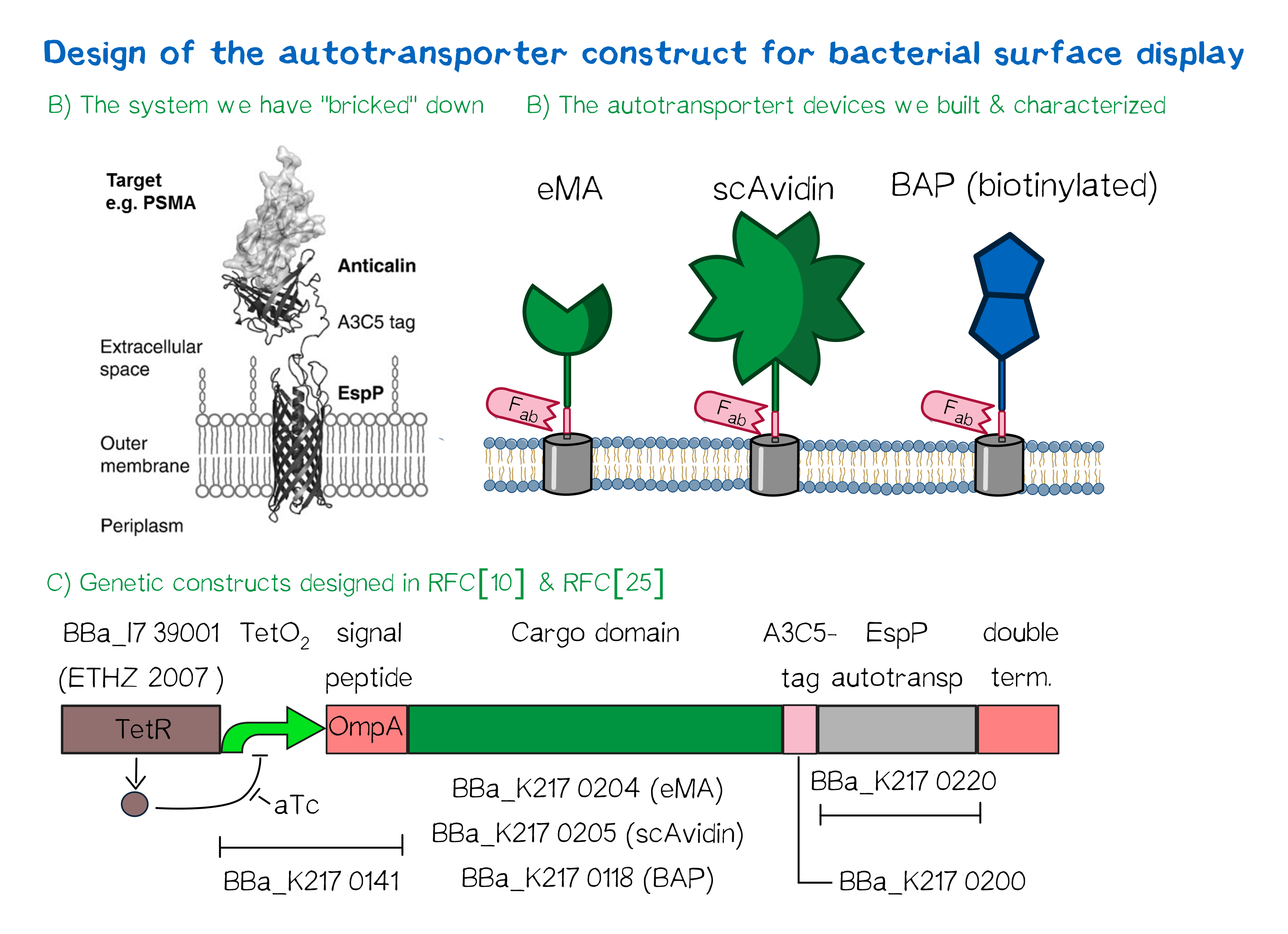 Figure 5
Figure 5