(→Week 5 (June 13th - June 19th)) |
(→Week 6 (June 20th - June 26th)) |
||
| Line 46: | Line 46: | ||
<div class="labbook"> | <div class="labbook"> | ||
</div> | </div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=Week 7 (June 27th - July 3rd)= | =Week 7 (June 27th - July 3rd)= | ||
Revision as of 16:11, 29 June 2016
Labjournal
Week 7 (June 27th - July 3rd)
Monday, June 27th
Sequencing of P67 (EGFR-Signalpeptid)
Investigator: Niklas
Procedure:
Sequencing batch was prepared after manufacturer's protocol. (15 µl of plasmid DNA (100 ng) and 2 µl sequencing primer (VF2))
FR11326586
Repetition of Quick-Change PCR of P3 (pASK + SAm1)
Investigator: Luisa
Procedure:
- The QC-PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 1,25 µl | Primer O21 |
| 1,25 µl | Primer O22 |
| 1 µl | dNTP-mix |
| 5 µl | Pfu-Ultra-II reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Pfu-Ultra-II Polymerase |
| 40,5 µl | ddH2O |
- Digestion of PCR-Product with DpnI for 1h at 37°C.
- Transformation of 10µl into component E.coli XL-1-blue, according to SOP (1h incubation at 37°C necessary despite AmpR).
PCR of Genesynthesis 3 and 4
Investigator: Luisa
Aim of Experiment: Amplification of Genesynthesis 3 (contains BAP and IGKappa) and 4 (contains A3C5-tag and BM40)
Procedure:
- The PCR was performed according the SOP.
- Reaction Mix:
| volume | reagent |
| 2,5 µl | Primer VF2 |
| 2,5 µl | Primer VR2 |
| 1 µl | dNTP-mix |
| 10 µl | Q5 Polymerase reaction buffer |
| 1 µl | template DNA (1:10 dilution of p3) |
| 0,5 µl | Q5-Polymerase |
| 18 µl | ddH2O |
- Setup: iGEM_standard (Promega-cycler)
| temperature | time |
| 98°C | 2min |
| 98°C | 10sec |
| 66°C | 30sec |
| 72°C | 30sec |
| 72°C | 2min |
| 4°C | hold |
- the batches were then purified using the Quiagen PCR-Purification Kit.
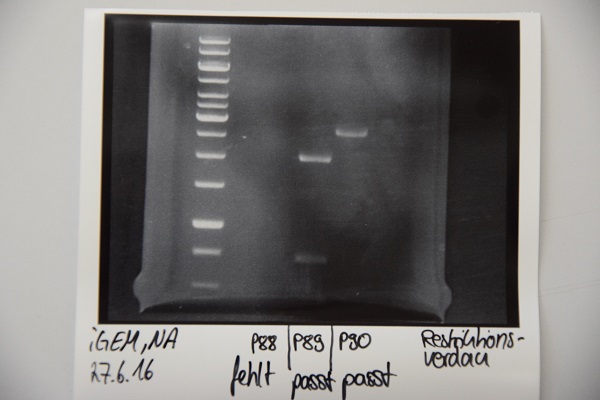
Analytical digestion and gelelectrophoresis of P88 , P89 and P90
Investigator: Niklas
Aim of experiment: Analytical digestion and gelelectrophoresis of P88 (pASK75 + Streptactin, former F58), P89 (CMV + CD4, former F65) and P90 (CMV + EGFR-signal-peptide, former F66)
Procedure:
- Batches for analytical digestions:
P88: EcoRI
P89: EcoRI and PstI
P90: EcoRI
| volume | reagent |
| 5,8/2,3/1,8 µl | Plasmid DNA (P88/P89/P90) |
| 1 µl | CutSmart buffer (10x) |
| 0.5 µl | EcoRI-HF(10 U/µl)/ PstI |
| required amount for total volume of 10 µl | ddH2O |
Ligation of F67 and F71, Transformation of E. coli XL1 blue afterwards
Investigator: Niklas
Aim of the experiment: Ligation of F67 (BirA) and F71 (empty pSB1C3), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 2,4 µl | Vektor |
| 7,6 µl | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| 7 µl | ddH2O |
| =20 µl | TOTAL |
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 750 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the cell-culture shaker.
- next step: analytic digestion of transformation was successful
Digestion of PCR on genesynthesis 3 and 4, and pSB1C3
Investigator: Luisa
Aim of experiment: Division of Leptin, IGKappa, A3C5, BM40 and BAP using SapI, HindIII, XbaI, AgeI for both batches.
Procedure:
- Batches for analytical digestions:
| volume | reagent |
| 1 µl each | enzyme (SapI, HindIII, XbaI, AgeI) |
| 5 µl | CutSmart buffer (10x) |
| 41 µl | DNA (purified PCR-products of GSY3 and 4) |
- Additionally 10µg of the vector P74 was digested with XbaI and AgeI in 100µl batch (2µl of each enzyme, 10µl of Cut-Smart buffer). Digestion was performed over night and purified via gelelectrophoresis and gelextraction according to the manufacturer's protocoll. --> Now labeled F80.
Analytical digestion and gelelectrophoresis of P80 , P78 and P85
Investigator: Julian
Aim of experiment: Analytical digestion and gelelectrophoresis of P80 (mRuby3), P78 (NanoLuc) and P85(Strep-Tag)
Procedure:
- Batches for analytical digestions:
- P80 and P78: EcoRI and AgeI
- P85: EcoRI and NgoMIV
| volume | reagent |
| 10 µl | Plasmid DNA |
| 32 µl | ddH2O |
| 5 µl | CutSmart buffer (10x) |
| 1.5 µl | each enzyme(10 U/µl)/ PstI |
| 50 µl | TOTAL |
Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3 with Strep-Tag) and Transformation into E. coli XL1 blue
Investigator: Luisa
Aim of the experiment: Ligation of F75 (mRuby) and F76 (NanoLuc) into F74 (pSB1C3+Strep-Tag), Transformation of E. coli XL1 blue afterwards.
Procedure:
| volume | reagent |
| 4,3µl(for F75), 8,2µl (for F76) | Vector |
| 12,7µl (F75), 8,8 (F76) | Insert |
| 2 µl | 10X DNA-Ligase-buffer |
| 1 µl | T4-Ligase |
| =20 µl | TOTAL |
- Ligation was incubated at RT for 1,5h.
- CaCl2 competent E. coli XL1-Blue cells were taken out of stock in -80 °C freezer and were gently thawed on ice.
- 7 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 15 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 950 µl LB-medium to each tube.
- Incubation for 1 hour at 37 °C
- The cell suspension was plated on Cam-plates (inclusive rescue plate) and incubated over night at 37 °C in the incubator.
Chemical biotinylation of BSA
Investigator: Niklas
Procedure:
- BSA was chemically biotinylated with a 20x and 40x molar excess:
- 10 ml of 100 mM borate buffer with 50 mM NaCl (pH 8.85)
- dissolve BSA (10 mg/ml)
- Add biotin-NHS-ester: 20,5 mg for 40x molar excess
- reaction over night
Tuesday, June 28th
</div>
Wednesday, June 29th
</div>
</div>