(→The receptors) |
(→Quantification of functionalized membrane proteins using flow cytometry) |
||
| Line 206: | Line 206: | ||
<br> | <br> | ||
| − | [[File:Muc16 Receptor_FACS_001.png |frameless| | + | [[File:Muc16 Receptor_FACS_001.png |frameless|left|550px]] |
=Immunochemical detecton of functionalized membrane protein using Western Blot= | =Immunochemical detecton of functionalized membrane protein using Western Blot= | ||
Revision as of 22:10, 18 October 2016
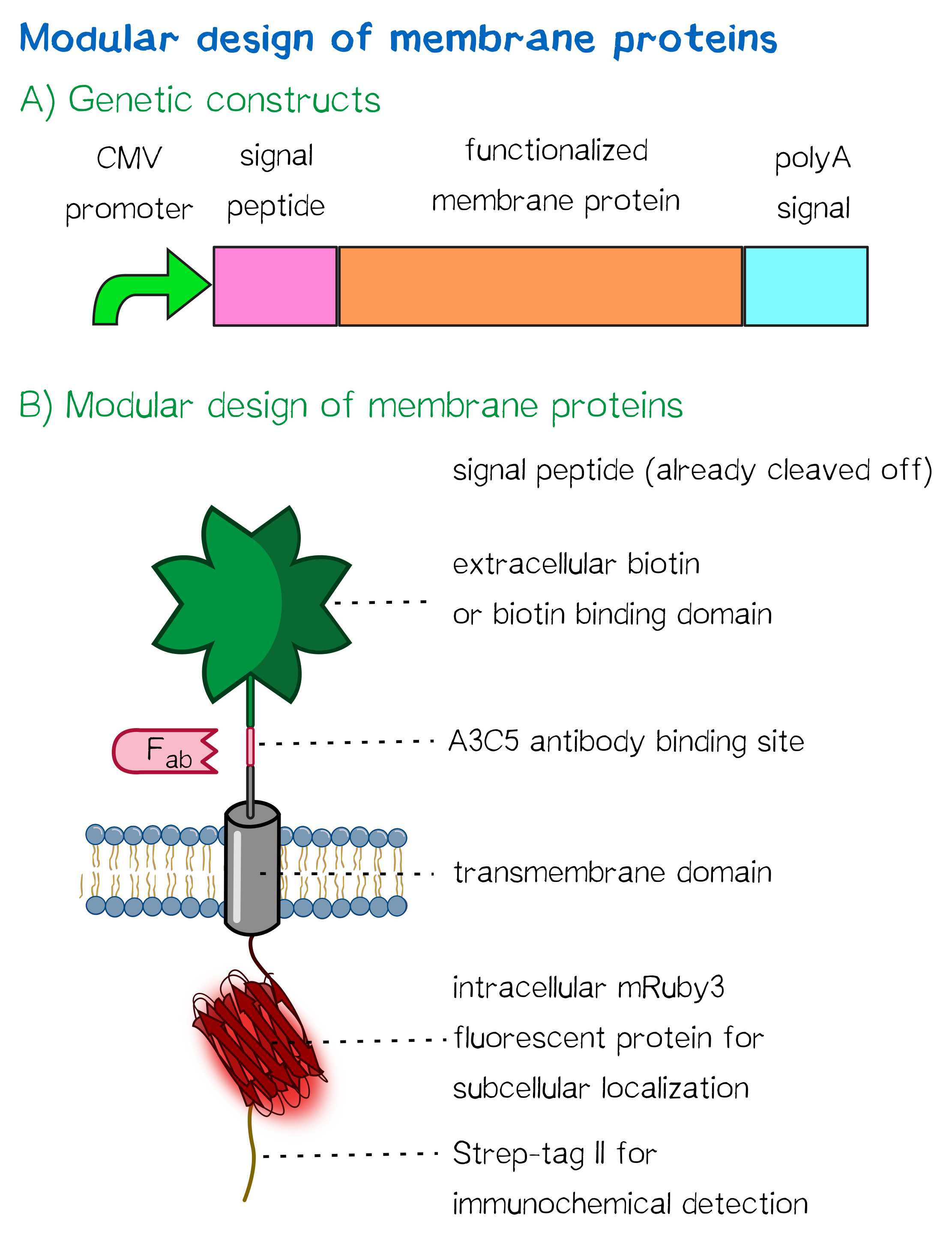
The receptors
Our receptors enable the polymerization of cells into three-dimensional cellular matrices during bioprinting. Therefore, our receptors present biotin or biotin-binding proteins on their surface, and - being co-injected with a biotinylated linker peptide into streptavidin solution - immediately 'glue' to each other by the means of synthetic biology, allowing printing of defined three-dimensional cellular structures.
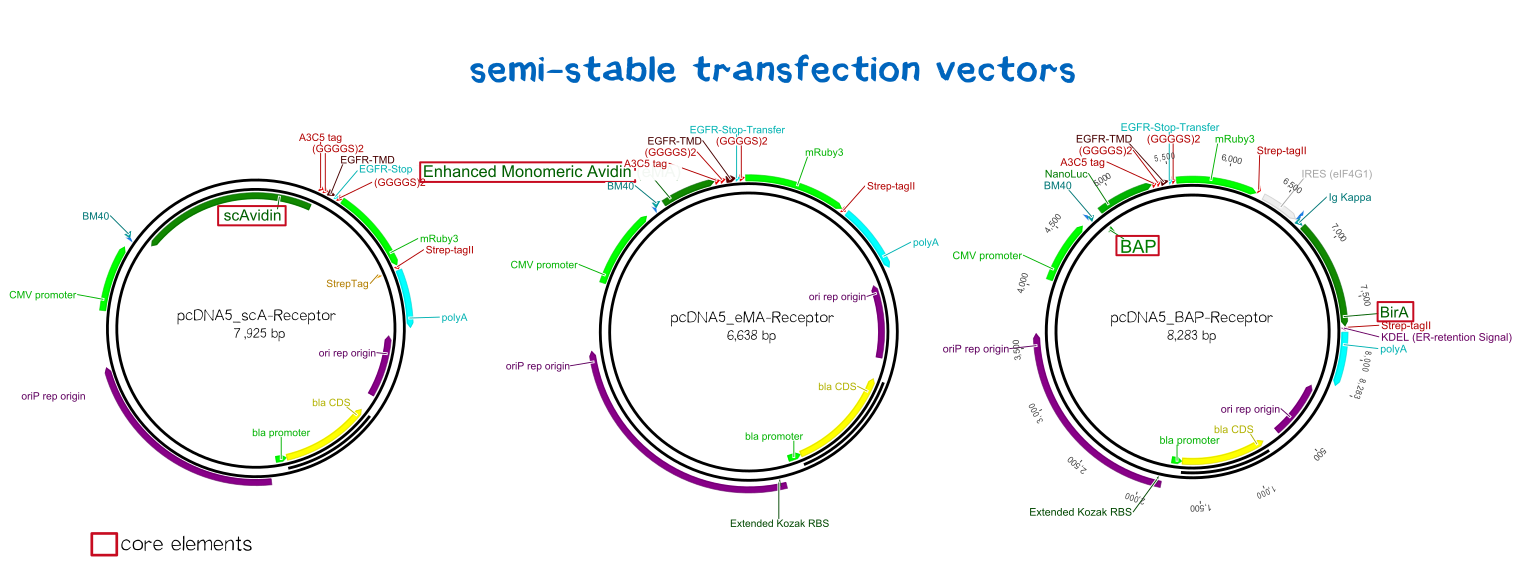
1) A receptor containing an extracellular biotin acceptor peptide that is endogenously biotinylated by a coexpressed biotin ligase (BirA) and thus presents biotin groups at the cell surface.
The biotin acceptor peptide (BAP) is a 15 amino acid peptide sequence originating from E. coli. [1] Being biotinylated by the biotin ligase BirA at a lysine residue within the recognition sequence, it mediates the functionality of the receptor by presenting biotin, allowing the interaction of the cell surface with streptavidin in the reservoir solution. The biotin ligase BirA is therefore encoded by the same vector as the receptor, with an internal ribosome entry site (IRES) allowing translation of two open reading frames (ORFs) from a single polycistronic plasmid. BirA is targeted to the ER via an Igκ signal peptide as well as an ER retention signal, allowing it to biotinylate translocating proteins possessing the recognition sequence.
2) Receptors presenting an extracellular, monomeric or single-chain avidin variant.
These avidin derivates, by design, allow the functional fusion of the otherwise tetrameric avidin molecule with our receptor. Two different variants were hereby used: The so-called enhanced monomeric avidin, a single subunit avidin that is able to bind biotin as a monomer, and single-chain avidin, which resembles the naturally occuring avidin tetramer, but has the subunits being connected via polypeptide linkers.
Both receptors generally mediate the same purpose: By interacting with streptavidin in the printing reservoir - either directly by presenting biotin (biotin acceptor-peptide) or indirectly via binding to a biotinylated linker (single chain-avidin variants), cells are being cross-linked due to the polyvalent binding of streptavidin molecules in the printing solution to several cellular biotin groups at once.
Prediction of signal peptide functionality
For determining a signal peptide suitable for our construct, we tested a total of three signal peptides - one taken from the parts registry, and two designed by ourselves.
- The EGFR signal peptide was taken from the iGEM parts registry ([http://parts.igem.org/Part:BBa_K157001:Design BBa_K157001]) and combined with a BioBrick encoding the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard.
- The BM40 and Igκ signal peptides were designed by us, providing additional spacing as well as a Kozak sequence to the 5'-UTR of the constructs and combined with a BioBrick encoding the CMV promoter sequence ([http://parts.igem.org/Part:BBa_K747096 BBa_K747096]) via the RFC10 cloning standard.
As shown below, the EGFR signal peptide taken from the parts registry is constructed in a way that the signal peptide ORF immediately follows the RFC10 cloning scar after the CMV promoter, thus resulting in a very short 5’ untranslated region (UTR) of the transcribed mRNA. The combination of the CMV promoter with the synthesized BM40 and Igκ signal peptides allows for a considerably longer 5’ UTR of the resulting mRNA - additionally allowing them to contain a full Kozak consensus sequence by design. The Kozak sequence is recognized by the ribosome as a translational start site; this element missing or deviating from the consensus sequence may considerably decrease translation efficiency.[2] For the EGFR signal peptide construct, a Kozak sequence is not present, as it would have to preceed the start codon ATG - a position which is occupied by the RFC10 cloning scar.
Since both the distance from the promoter to the open reading frame as well as the Kozak consensus sequence are considered crucial parameters for expression levels[3], BM40 and Igκ signal peptide constructs are likely to resut in increased expression levels compared to proteins containing the EGFR signal peptide from the parts registry.
As the quantitative functionality of signal peptides had to be determined before including one of these into the final receptor constructs, the three options (Igκ, BM40, EGFR) were tested via bioinformatic tools as well as a secretion assay of luciferase constructs.
Bioinformatic signal peptide analysis via the SignalP server
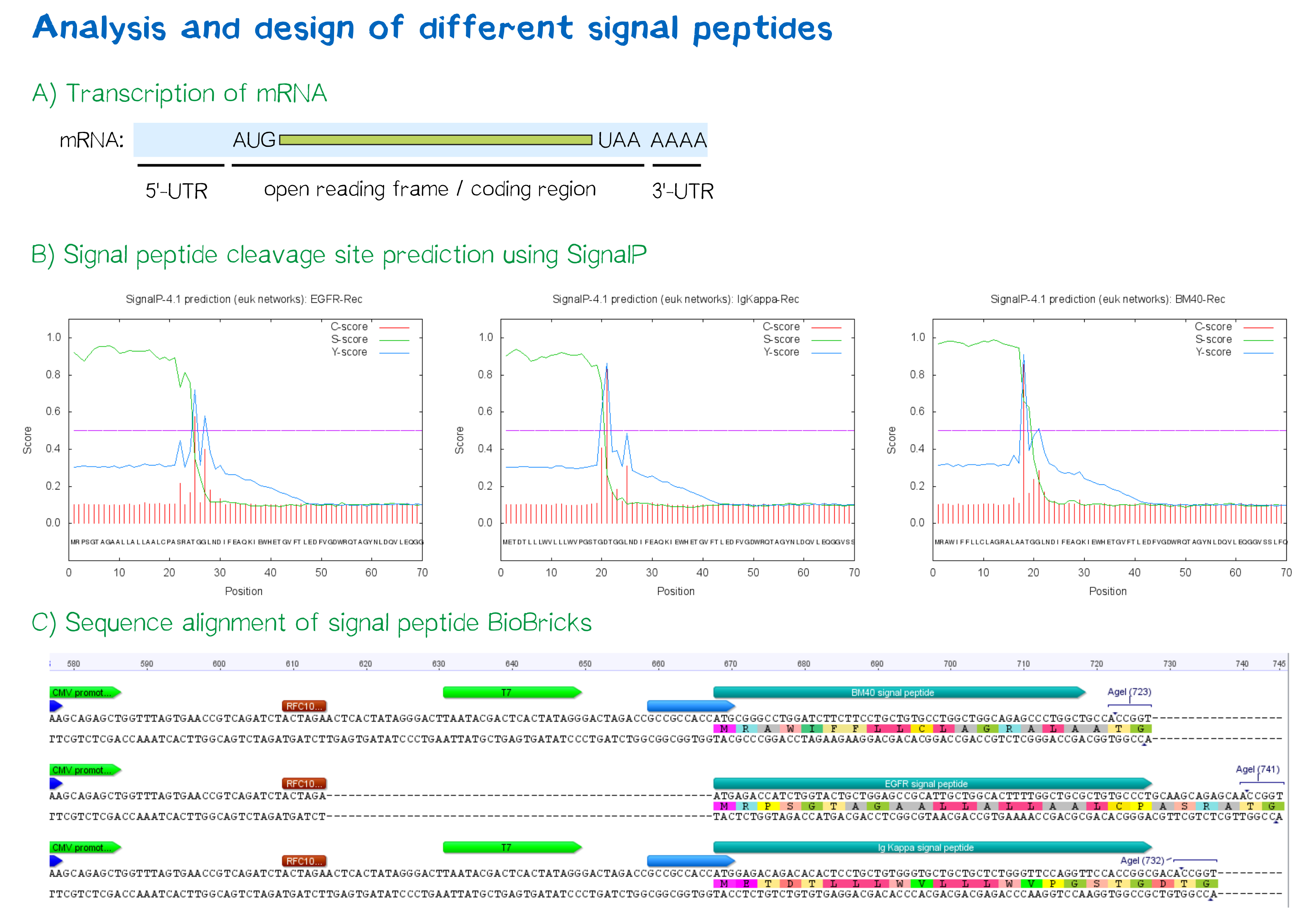
For all three signal peptides, the high S-score indicates high signal peptide functionality. For the Igκ signal peptide, a cleavage site between amino acid 20 and 21 within the signal peptide is predicted. For the BM40 signal peptide, a cleavage site between amino acid 17 and 18 is predicted, and for the EGFR signal peptide, a cleavage site between amino acid 24 and 25 is predicted.
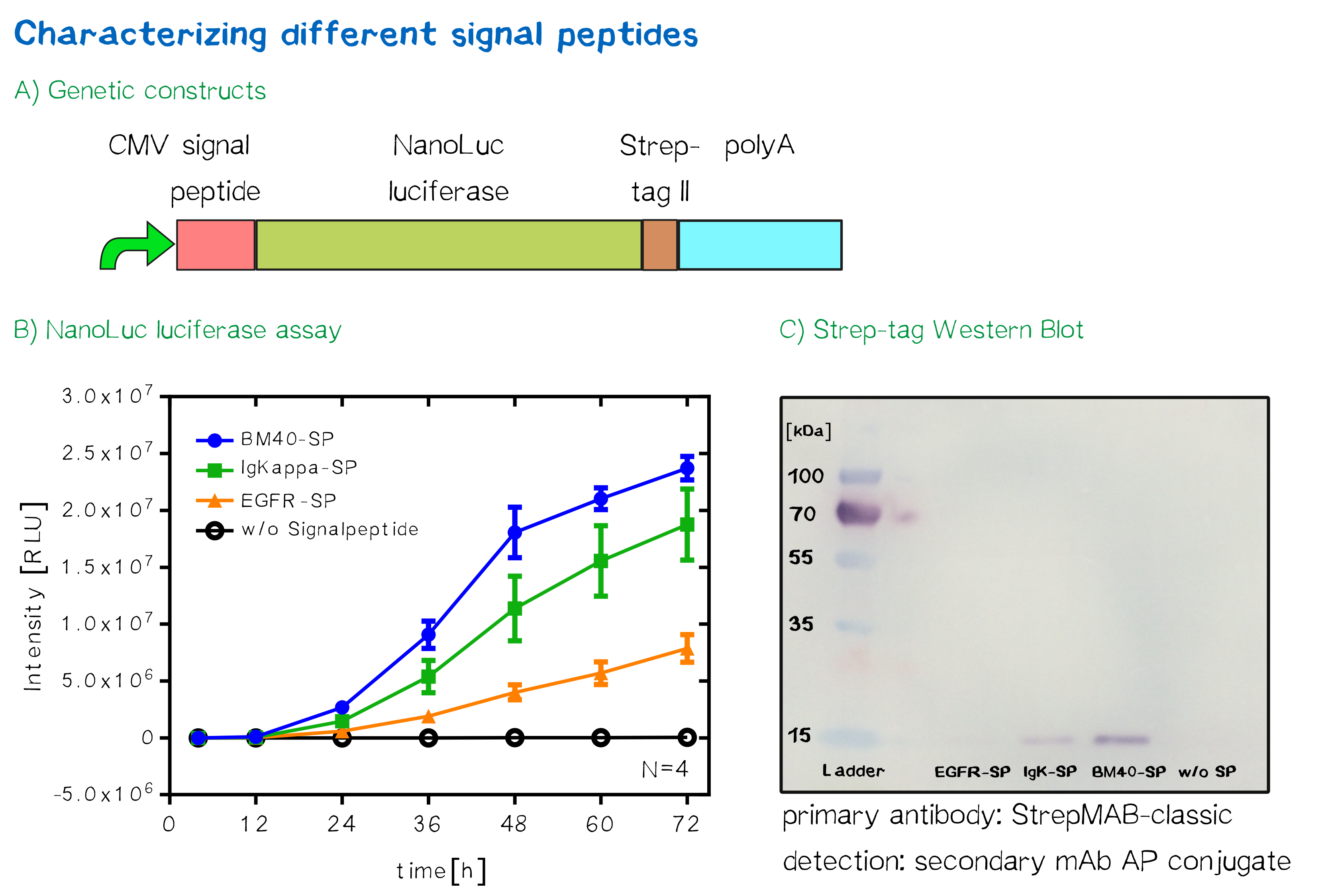
Quantification of signal-peptide mediated translocation via a secretion luciferase assay
For the quantification of signal peptide functionality via a luciferase assay, three constructs were created and tested - each containing one of three signal peptides (the EGFR signal peptide, the Igκ signal peptide or the BM40 signal peptide) and a nanoluciferase as well as a CMV promoter, a Strep-tag II for immunochemical detection and the hGH polyadenylation signal sequence. Not containing a transmembrane domain, the nanoluciferase fusion protein is being translocated into the ER and then secreted into the medium. Using a luciferase assay, one can quantifiy the amount of luminescence - and thus, proportionally, the amount of secreted luciferase - by measuring the conversion of luciferin into visible light and integrating it over a timespan of 5 s. Therefore, medium samples were taken every 12 h after transfection of cells and measured via the Promega NanoGlo luciferase assay system according to the manufacturer's instructions.
Other receptor elements: stability and detection
Apart from the functional parts that mediate the (strept)avidin-biotin interaction, other elements in the receptor were designed to make sure it is optimally translocated to the membrane, as stable as possible, and easily detectable.
EGFR transmembrane domain
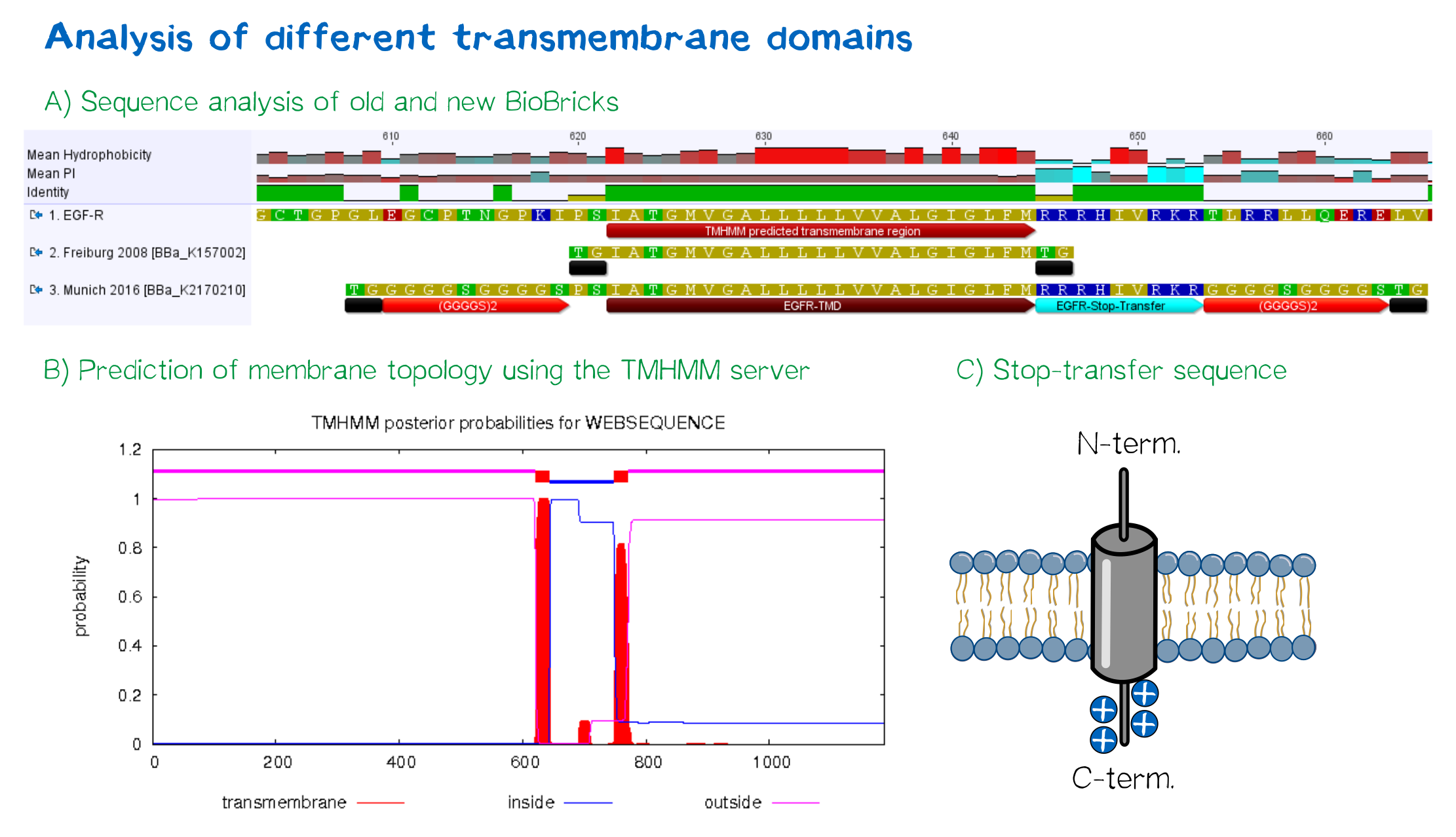
For anchoring in the membrane, we decided to use a type I membrane protein, which has a single membrane span with defined localization of N- and C-terminus out- and inside of the cell, respectively. The N-terminal transmembrane α-helix of human EGFR ([http://www.uniprot.org/uniprot/P00533 UniProt P00533], amino acids 622-653) was herefore chosen (see below the topology prediction of EGFR via the [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM 2.0 server for the prediction of transmembrane helices][5]). A stop-transfer sequence consisting of charged amino acids - being characteristic for type I membrane proteins - was added at the C-terminus, and the sequence was furthermore flanked by a (GGGGC)2-linker at the N- and C-terminus, respectively. The addition of a stop-transfer sequence as in the naturally occuring EGFR sequence, as well as the addition of flexible linkers, makes our EGFR-TMD an improvement over the BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K157002 BBa_K157002], which does not possess either of these elements.
Moreover, the receptor contains three functional elements for its detection: The intracellularly located red fluorescent protein mRuby 3 for detection of the receptor via fluorescence microscopy, an extracellular epitope domain for immunochemical detection via A3C5-antibody fragments and an intracellular Strep-tag II for the detection and purification via immunochemical methods.
The vector furthermore contains the poly-adenylation signal of human growth hormone (hGH) for functional polyadenylation of the transcribed mRNA. A description for the respective functional elements is given in the following sections.
A3C5 epitope tag
Antibodies have various areas of application in life science, including protein detection, pulldown experiments and even immunotherapy. Having originally been discovered in 1995 during an in vitro screening for antibodies against cytomegaloviral proteins[6], the A3C5 antibody is an established molecular tool for specific recognition of proteins. By tagging cell surface proteins with an epitope specifically recognized by A3C5 (being a peptide sequence of 11 amino acids), one is easily able to detect tagged proteins via immunofluorescence microscopy, FACS or one may purify them via pulldown experiments. Through the latter, one may also screen for in vivo interaction partners of the tagged protein.
mRuby 3
Having originally been engineered as a monomeric form of the red fluorescent protein eqFP611, mRuby variants are sine of the brightest red fluorescent proteins available. [7] With an excitation maximum at a wavelength of 558 nm and an emission wavelength maximum of 605 nm, the resulting stokes shift of 57 nm makes mRuby a good choice for fluorescent microscopy imaging and, thus, allows the visualization of the receptor in its cellular environment by being fused to the C-terminus of the transmembrane domain. mRuby is especially powerful for the fusion with receptors - not only due to its small size and high stability, but also due to the fact that, in comparison with eGFP, it appears up to 10 times brighter in membrane enviroments. The mRuby variant used here is mRuby 3, having been engineered for even more improved brightness and photostability.[8]
Strep-tag II
The Strep-tag II is an eight amino acid peptide sequence that specifically interacts with streptavidin and can thus be used for easy one-step purification of the receptor via immunochemical methods.[9]
Nanoluciferase
The concept of bioluminescence has a great meaning for several invertebrate species, allowing communication with other individuals, signaling receptiveness or deterring predators.[10] The most common enzyme responsible for the creation of bioluminescence are the luciferases (from the latin words ’lux’ and ’ferre’, meaning ’light-carrier’). These enzymes, among others found in fireflies and deep-sea shrimp, commonly consume a substrate called luciferin as well as energy in the form of ATP or reduction equivalents in order to create light-emission through oxidation. The underlying mechanism hereby relies on the creation of an instable, highly excited intermediate under the consumption of energy. This high-energy intermediate then spontaneously falls back into a state of lower energy, emitting the energetical difference as visible light. While mainly being used as a folded, stabilizing element at the N-terminus for the construct containing the biotin acceptor peptide, the nanoluciferase could also be used for advanced binding studies via luciferase assays.[11]

Luciferases have also found their way into biotechnological applications.[12] The simplistic concept of creating visible (and thus easily measurable) light makes luciferases ideal reporters for the expression of secretable proteins via a corresponding assay. This project also made use of a luciferase for the determination of expression levels of the BAP surface receptor while providing a stable and folded domain at the N-terminus, as well as having been used for the choice of signal peptide. The monomeric luciferase used in this project, the so-called ’NanoLuc™'[13] (due to terms of simplicity referred to as ’nanoluciferase’ in the following), can be fused to other proteins in order to make their expression visible and quantifiable. This engineered luciferase emits a steady, easily detectable glow when exposed to its substrate, while being only 19 kDa large and being brighter, more specific and steadier than other luciferases commonly found in nature. Using luciferases for the quantification of expression levels offers several advantages over fluorescent proteins, including higher sensitivity and smaller size.
CMV promoter
Last but not least, high expression levels of the receptor, so that a high avidity for the interaction may be reached. The CMV promoter (taken from the parts registry, [http://parts.igem.org/Part:BBa_K74709 BBa_K74709]) therefore enables constitutively high expression of the receptor - and, in the case of the BAP-construct, the biotin ligase BirA.
BioBrick-compatible vectors for mammalian cells: transient transfection & stable integration
Semi-stable transfection of the receptors in human cells
For the first trials of testing our constructs in human cells, we chose the MEXi™ cell line for semi-stable transfection. MEXi™ is a suspension HEK293E cell line by iba lifesciences that encodes the viral protein EBNA1 in it's genome. EBNA1, being a protein derived from Eppstein-Barr-Virus that mediates the viruses' replication initiation, gene regulation and maintenance of its DNA in the host cell. The second part of the MEXi™ system is made up by the pDSG-IBA vector that carries the specific origin of replication that is recognized by EBNA1. EBNA1 binds to this origin, and recruits the replication apparatus as well as inducing the maintenance of the inserted DNA in the host's nucleus, leading to the episomal maintenance and co-replication of the plasmid during every division cycle.[14]
Thus, we cloned our receptor constructs into the pDSG-IBA vector and then transfected the MEXi™ cells using the PEI transfection method (see methods). Polyethylene imin (PEI) as a cationic polymer forms complexes with the negatively charged DNA. These complexes are taken up by the cell via endocytosis and carried to the nucleus. [15]
- Abbildung semistabile transfektion schematisch
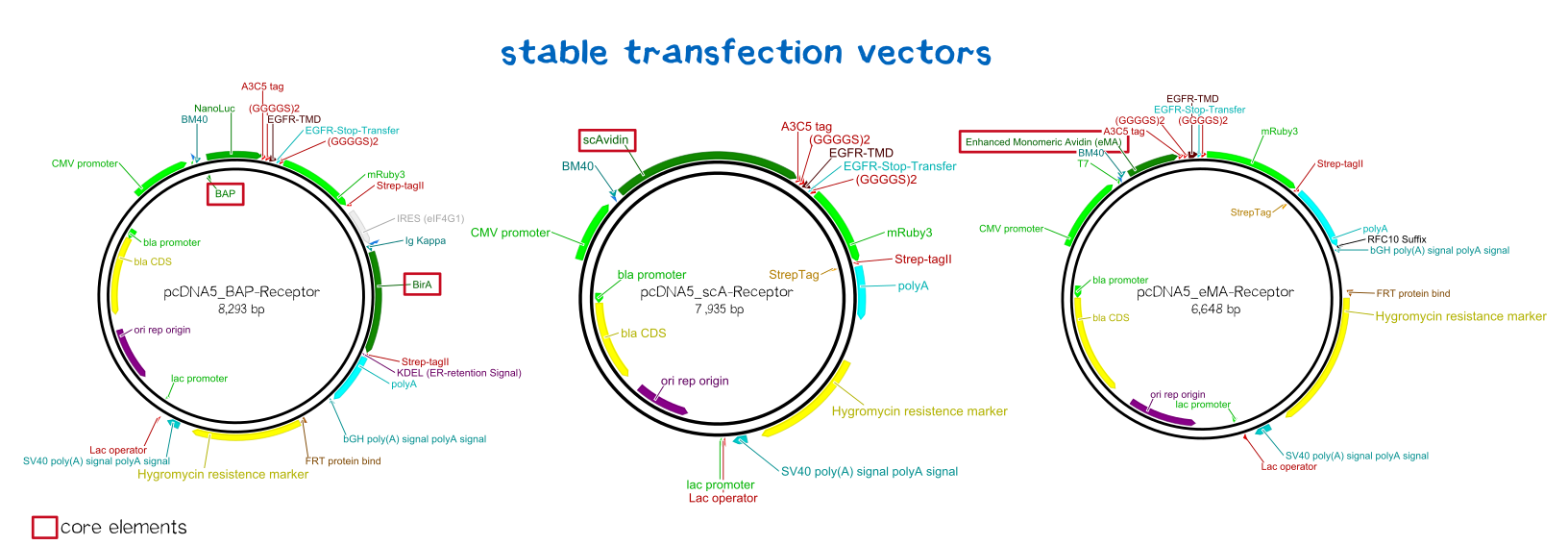
An everlasting bond: creating a stable cell line
Printing tissues - or even only parts of organs - requires billions and bilions of cells. An average liver, for example, weighs about three pounds and hosts ~108 cells per gram tissue. [16] [17]
Since transiently and even semi-stably transfected cells do not propagate the transfected DNA indefinitely, repeated transfection of cells would be unwise due to the high consumption of time and ressources. Therefore, we created three stable cell lines that chromosomally encode the respective receptor constructs. For this purpose we used the FlpIn™-System by invitrogen. The three compound system consists of HEK-TRex™-293 cells that harbour one chromosomal Flp recombination target (FRT) site, a Flp recombinase expression plasmid and the plasmid encoding the receptor as well as another FRT site for stable integration.[18] The detailed principle of the intergration is described in the figure below.
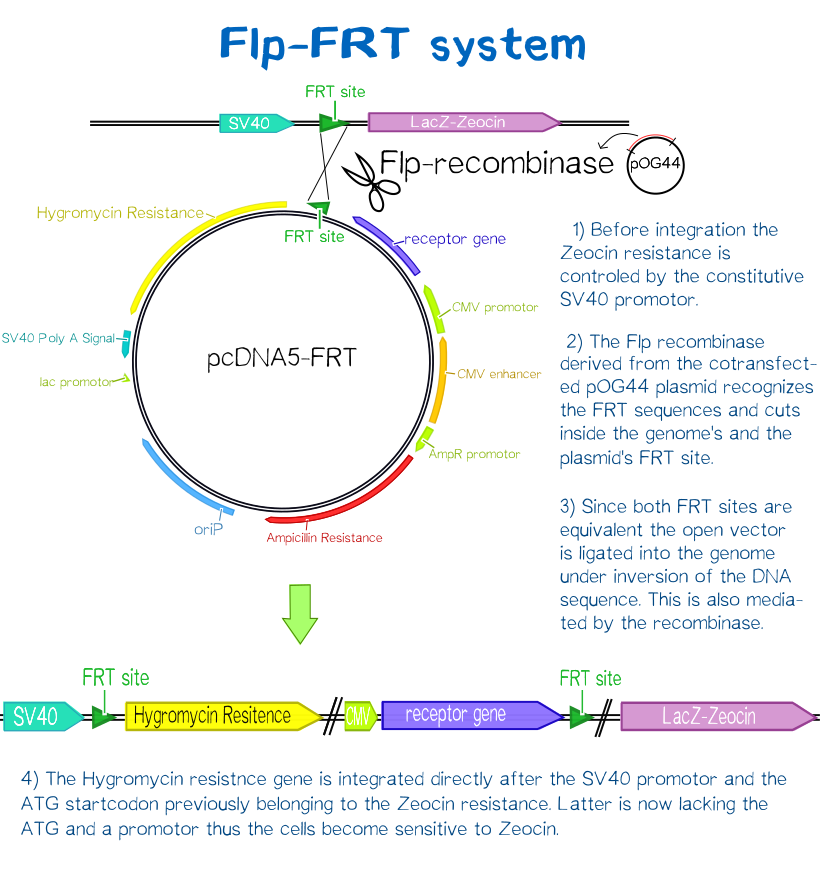
Thus the transfected cells can be easily selected via hygromycin B. To ensure that the resistence does not come from non-intergrated plasmid, this process takes several weeks and divison cycles.
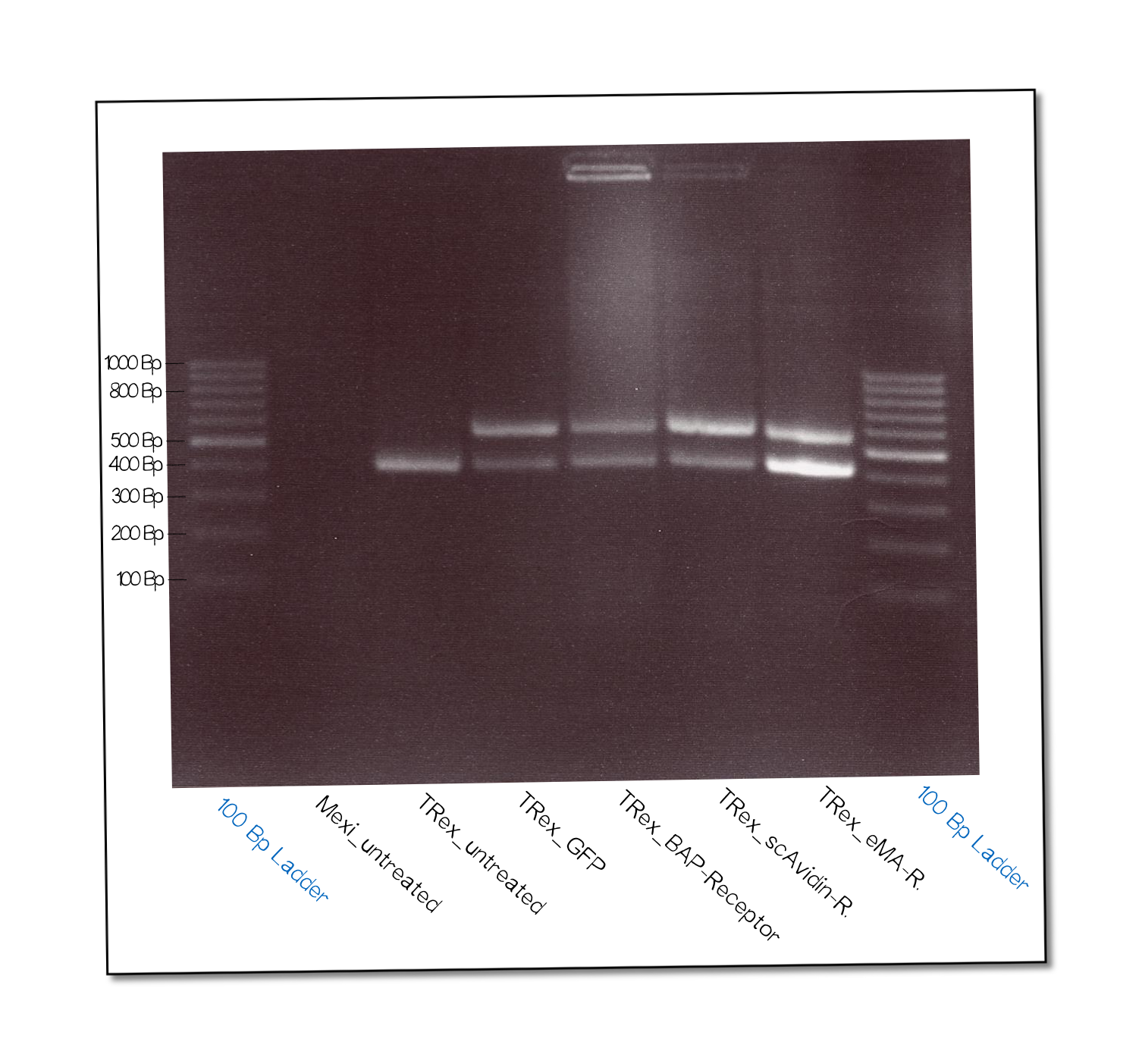
The succesful integration of the pcDNA plasmids was verified by genome-PCR (see methods). Therefor we designed three primers: One that binds to the SV40 promotor and part of the following FRT site, one that binds part the hygromycin restistance gene and another one for the lacZ-Zeocin sequence. In the event of stable integration the size of the amplicons will be around 550 bp, according to the distance between the SV40 and the hygromycine resistance gene. Wheareas in case of no intergration the amplicon size will be around 400 since this corresponds to the distance between the SV40 and the lacZ-Zeocin sequences. The results are shown in figure on the left. The lower band visible in the samples 2-5 leads us to the conslusion that despite hygromycine selection the cells are not yet a monoculture, something we also observed in the flow cytometry analysis further discussed below.
- Abbildung mit stabiler PCR-Primern, Zeocin-Test, Bildern vor und nach Transfektion
Subcellular protein localization using fluorescence microscopy
For bestter traceabilty we supplied all of our receptor constructs with reporter genes like mRuby and/or Nanoluciferase.
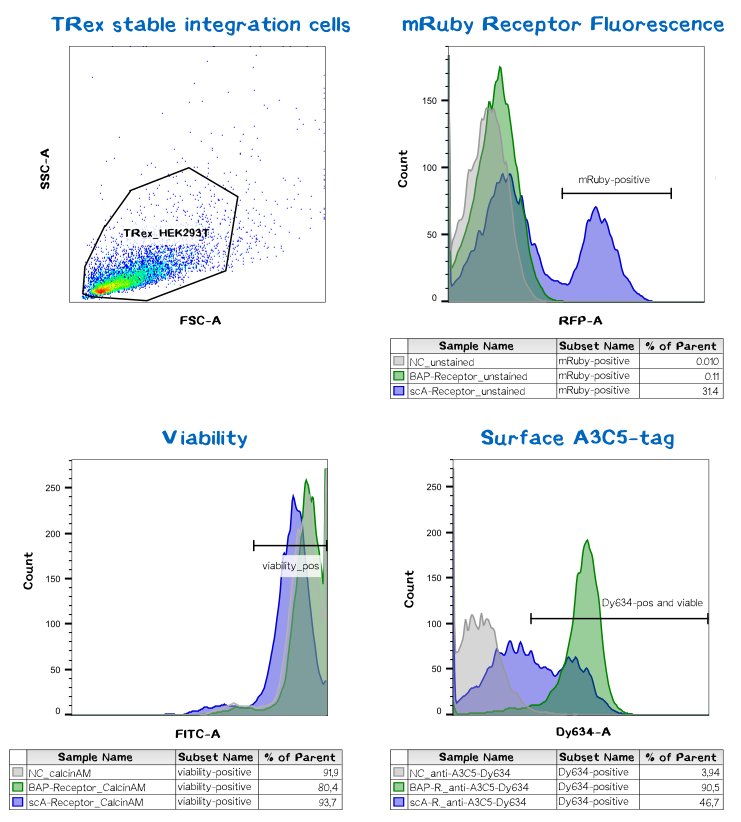
Quantification of functionalized membrane proteins using flow cytometry
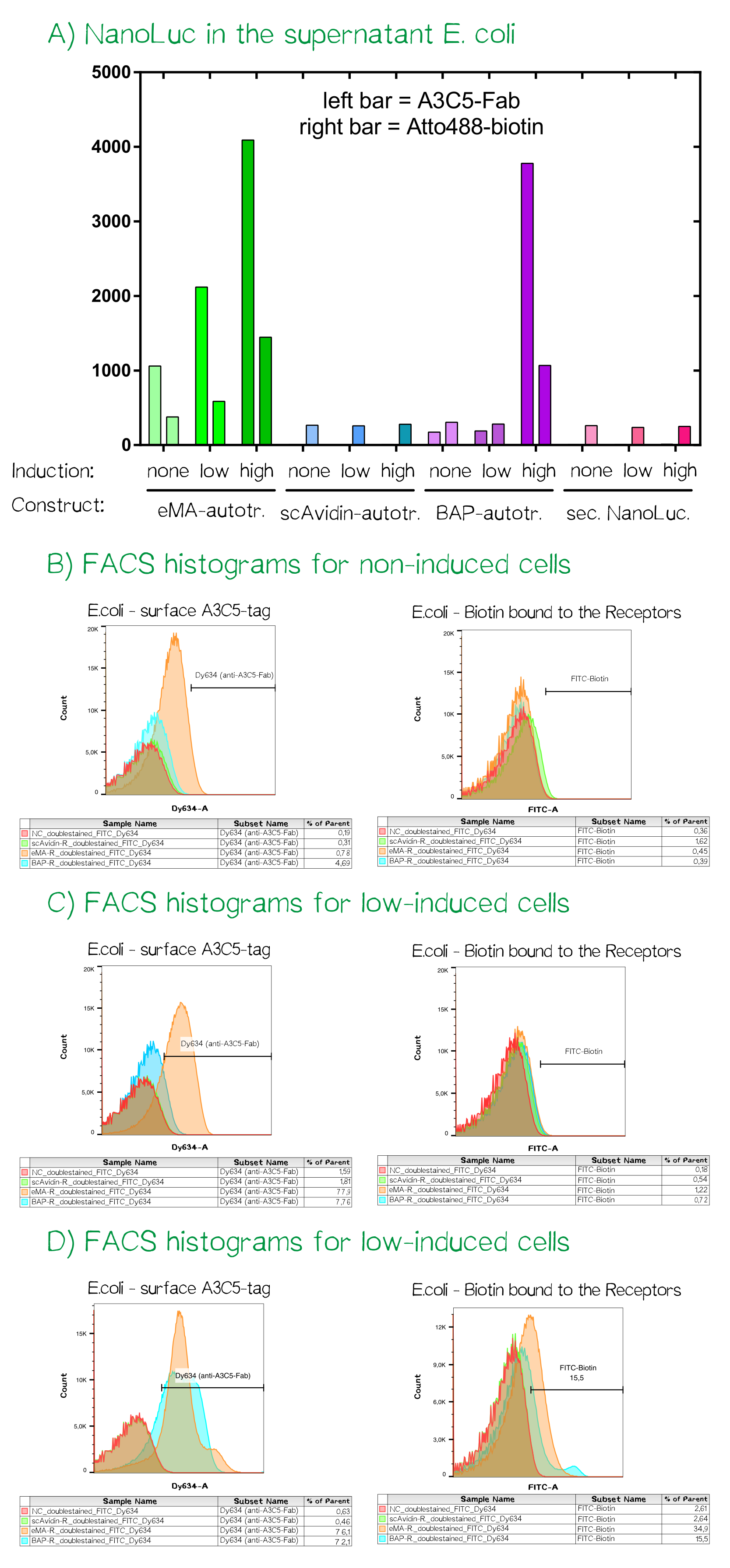
To make sure the receptor is properly presented on the surface of the cells and to quantify the yield of stably transfected cells we used flow cytometry analysis. In order to check the viability of the cells the dye calceinAM was used. This aromatic calcein derivative passes the cell membrane. In the cytoplasm the acetoxymethoxyl (AM) group is cleaved of by esterases what enables the substance to complex Ca2+ ions leading to a bright green fluorescence. The cells where additionally incubated with anti-A3C5-Fab labeled with Dy634, a fluorescent dye emitting light of 658 nm wavelegth. The intrinsic fluorescence made up by mRuby could - as visible in the graph below - only be detected in the scAvidin receptor cells and not in the BAP-Receptor cells. This was not further surprisingsince the cells didn't show hardly to no fluorescence under the microscope as well. But interestingly we got a positive signal for A3C5 (anti-A3C5-Dy634) meaning that the protein must be expressed and as well be located in the membrane.
Immunochemical detecton of functionalized membrane protein using Western Blot
Another way to verify the succesful transfection and protein expression is given by the two tags (A3C5 and Strep-Tag) that are included in the receptor design, which allow detection via western blot analysis.
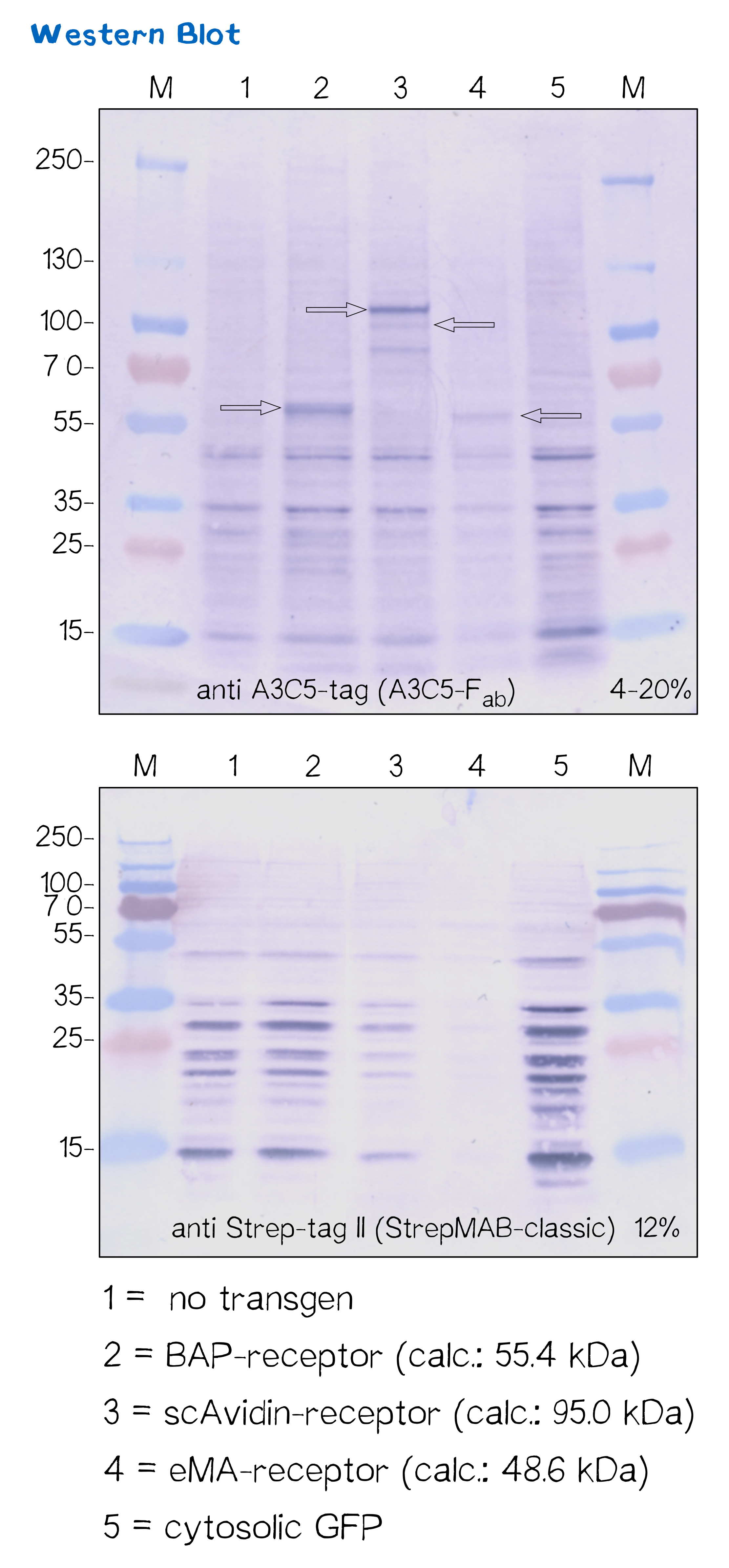
After transfecting mammalian cells with the receptor constructs and selecting them for 2 weeks on hygromycin, we lysed the cells and checked for the presence of Strep- and A3C5-tag via western blot using primary antibodies against the respective tags und secondary alkaline phosphatase coupled antibodies (see methods). As the figure shows, the Strep-tag wasn't detectable via WB - however, the A3C5-tag-WB shows significant singular bands at the size of our proteins.
This data corresponds with the flow cytometry results again showing that the BAP receptor is also present in the non-fluorescent cells.
| Receptor | BioBrick | Amino acids | Moelcular mass [Da] |
| BAP-Receptor | [http://parts.igem.org/Part:BBa_K2170000 BBa_K2170000] | 507 | 55 440 |
| eMA-Receptor | [http://parts.igem.org/Part:BBa_K2170001 BBa_K2170001] | 448 | 48 618 |
| scAvidin-Receptor | [http://parts.igem.org/Part:BBa_K2170002 BBa_K2170002] | 877 | 95 028 |
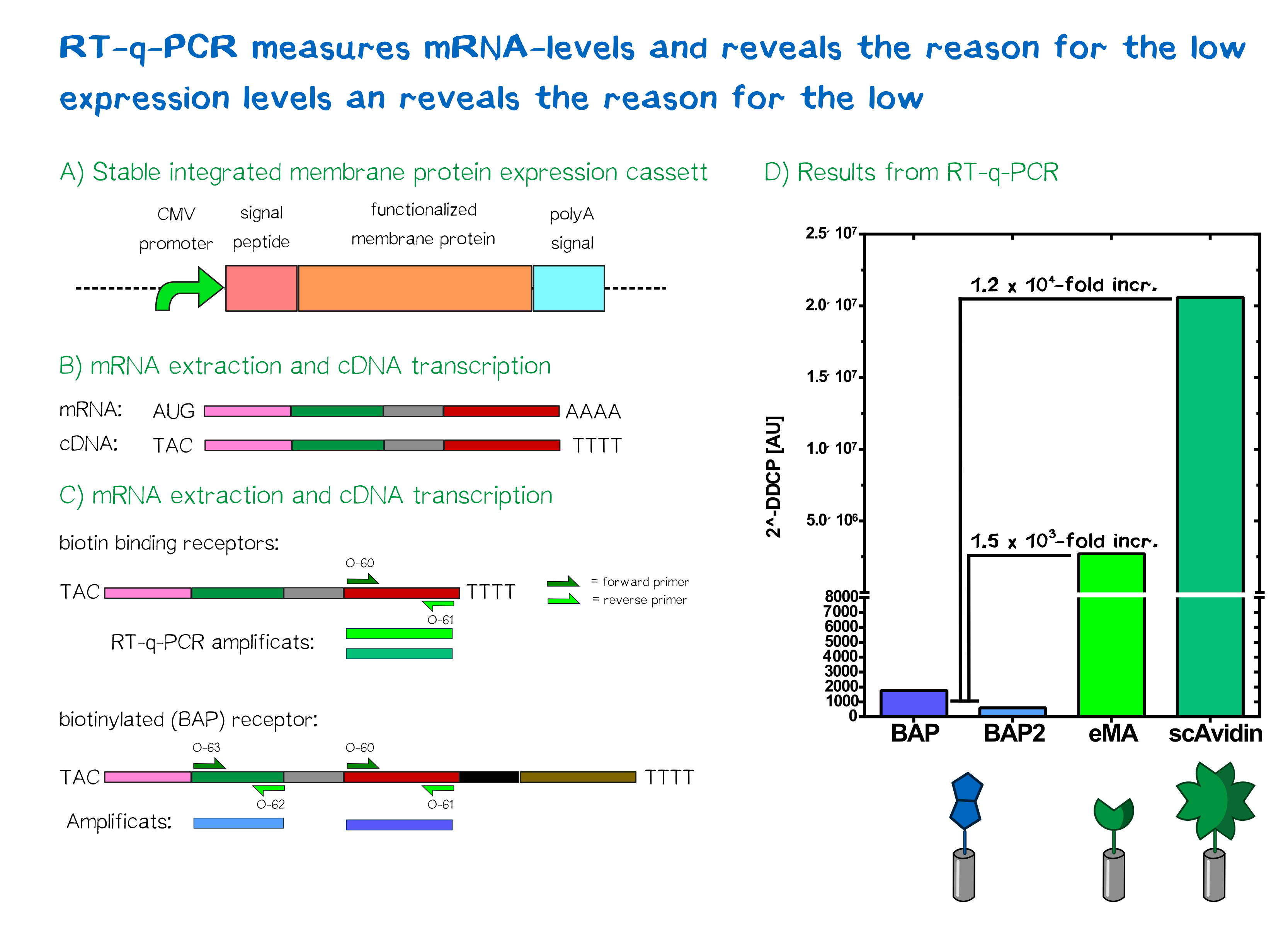
Quantification of mRNA expression levels by RT-q-PCR
The characterization of our transfected mammalian cells showed that the two receptors containing an avidin variant were expressed and translocated to the outer cell membrane. The biotin-presenting receptor containing the biotin-acceptor peptide, though, showed a very low fluorescence signal. Therefore, the success of the stable integration was tested by PCR of the genomic integration, which lead to the conclusion that the construct was transfected and integrated sucessfully, hinting at a problem downstream of the integration. In order to understand where the source of this problem was, we took a closer look at the expression levels of the different receptors by analyzing mRNA levels in comparison to each other and to the housekeeping gene (HGK) GAPDH.
For a better comparibility, we used the same amount of cells for each receptor and not only reviewed cells with stably integrated but also cells with the episomal stable receptor constucts. After extraction of the total mRNA from the cells the quality and integrity was measured by Bioanalyzer. All samples showed a RNA integrity number (RIN) above 9 which means that the total mRNA is pure and does not have any contamination. During the first step of RT-qPCR the total mRNA in converted to cDNA by a reverse transcriptase, which is then used in the actual quantitative PCR to quantify the amount of a specific mRNA was produced by the cells. For this reason two primer pairs were designed. One of them was designed to bind the exact same region in all of the different receptor mRNAs and amplify the same part of the receptor. Contrary to that the second pair was supposed to bind only in the biotin presenting receptor and amplify a part that is unique to this receptor. By this method we wanted to observe if only one part of the mRNA is unstable or if the general mRNA level of this receptor is decreased.
For the evaluation of the raw data we used the ΔΔCP method[20] in which the crossing points (CP values) of a gene of interest and a of reference gene are compared, to get an idea of the relative amounts of the mRNA of interest.
In the following you can see the results of our experiments. We compared the expression levels of Trex which were transfected with our different receptor constructs and with non-transfected (NT) Trexs.
As can be seen the ΔΔCP values of the mRNA of the eMA and scAvidin constructs are with a 1,2 x 106 fold increase for scAvidin and 1.5 x 103 fold increase for eMA, significantly higher than those of the BAP construct. The analysis of the different primer pairs in the two regions of the BAP construct also showed similar values. A difference in CP value is tatamount to the relative amount of the analysed mRNA in the cell.
Together with the result that the integration of the constuct into the genome was successful we can conclude that our first assumption, that the missing RFP signal in cells, which were transfected with the BAP construct, is caused by a reduced mRNA stability, is correct.
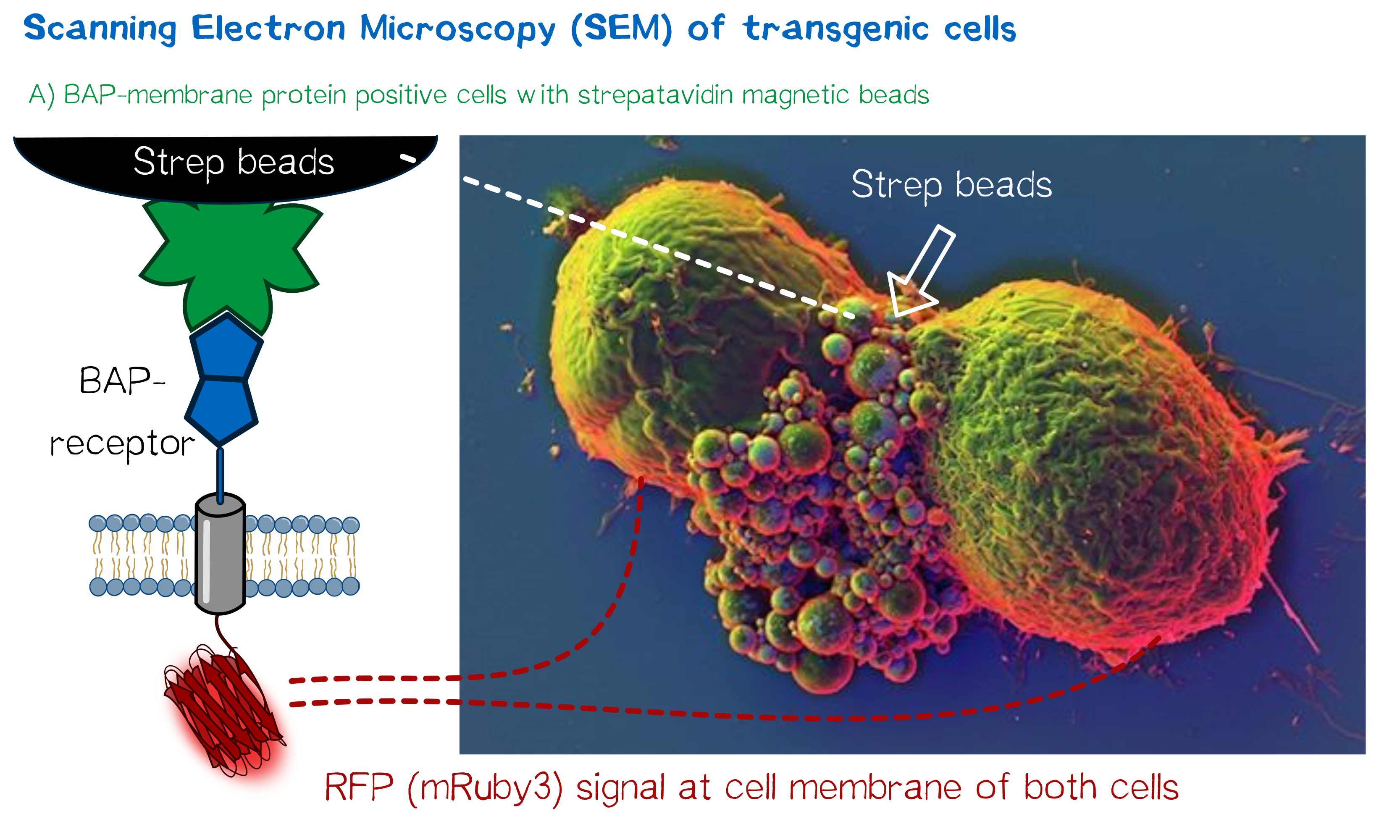
High resolution imaging using scanning electron microscopy (SEM)
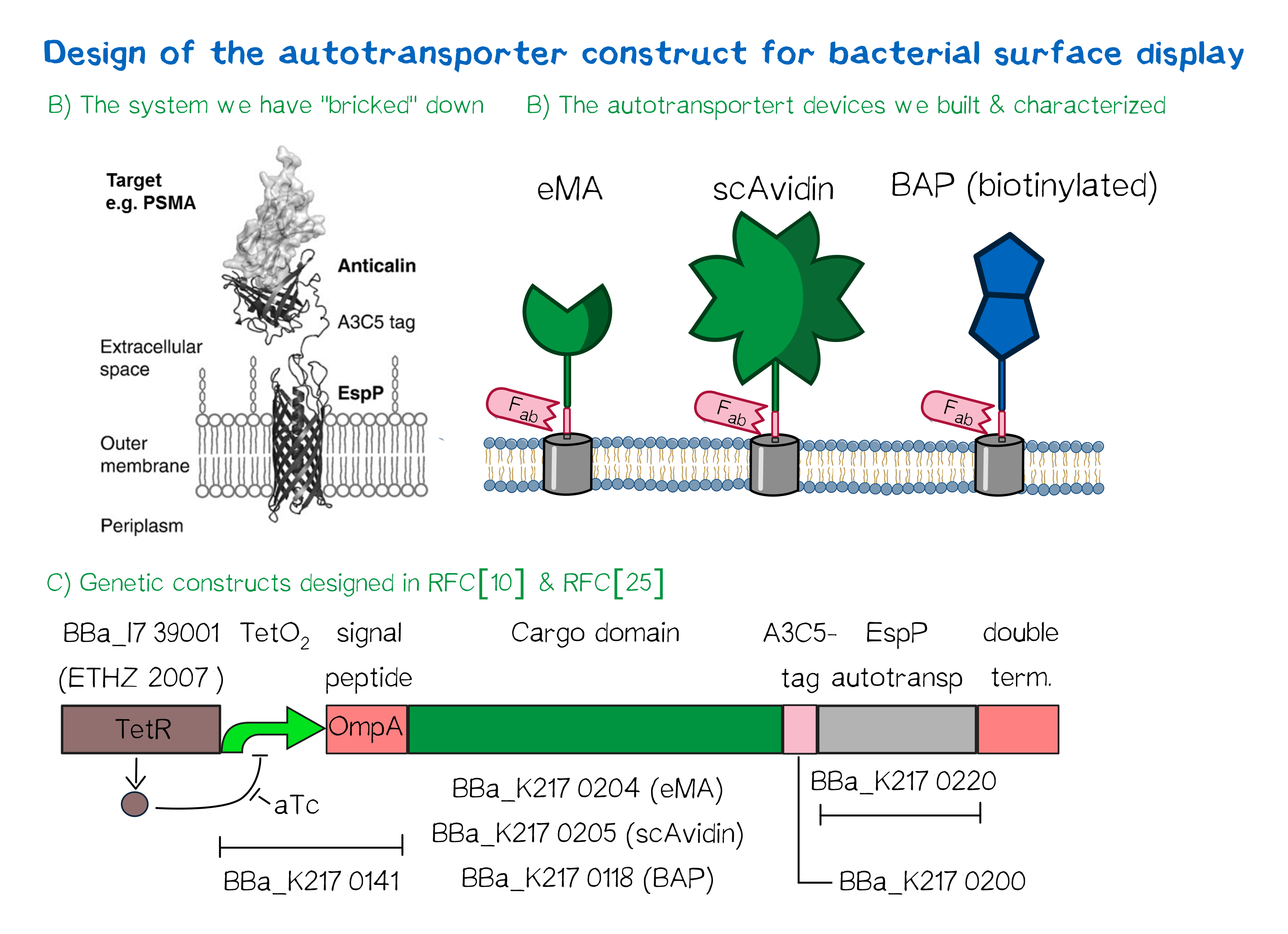
Design of an autotransporter construct for bacterial surface display
Besides the design of biotinylated and biotin-binding receptors for eukaryotic cells we also wanted to apply our biotINK approach for prokaryotic cells. For this purpose we designed an autotransporter device that is able to present biotinylated or biotin-binding protein domains on the surface of E. coli. These constructs for the bacterial surface display are well known in the field of protein engineering of therapeutic proteins, such as antibodies. Here they are used to screen libraries of different antibodies concerning their affinity towards a given therapeutic target. We based our design on the autotransporter EspP that has been developed in the Lab of Prof. Skerra [21].
This system works very well (see Fig. A[22]) and we "bricked" it down into RFC[10] and RFC[25] BioBricks. At first we tested different inducible bacterial promoter systems (e.g. "TetR repressed GFP"; BBa_K577893) that are available in the parts registry, but when we saw that two of the available systems did not work in our own hands, we decided to built the system on our own. We used the Tet-Repressor generator that was available on the distribution plates (BBA_I739001) as we believe that it is very important for the standardization in SynBio to use available parts even if it is more work to replace genetic elements that are already in place. Downstream of this repressor generator we designed and fused a BioBrick (BBa_K2170141) that encodes a double Tet-Operator (Tet-Repressor binding site together with a bacterial promoter and an signal peptide of the outer membrane protein A (OmpA[23]) from E. coli that has a 3' RFC[25] restriction site to allow the protein fusion of proteins that are secretet into the bacterial periplasm when expressed with this BioBrick. As a proteinfusion we then assembled a protein cargo domain that is generally exchangeable for any protein that is so small in size that it can be transported through the outer membrane by the EspP autotransporter. In our project we used the same extracellular domains as for the eukaryotic receptors: enhanced monomeric Avidin (eMA; BBa_K2170204), single-chain Avidin (scAvidin; BBa_K2170205) and a composite part composed of a N-terminal Biotinylation acceptor peptide (BAP) and a C-terminal NanoLuciferase (together shown as BAP; ; BBa_K2170118, see Fig. B). Further downstream our design features a A3C5-epitope tag that can be recognized by a high affinity Fab-fragment and is generally used to detect and quantify the surface display of the cargo domain[24]. Further downstream of this affinity tag we fused the BioBrick for the EspP autotransporter from E. coli[25][26] which is then followed by a bacterial terminator. For the termination we again chose a well working terminator from the distribution plate ("double terminator"; BBa_B0010-BBa_B0012).
But this was just the DNA-part. What is expected to happens on a protein level? The construct constantly produces Tet-repressor that binds to thet Tet-Operator and repressed the promoter activity of the autotransporter gene. As soon as the inducer anhydrotetracycline (aTc) is added to the culture, it binds to the Tet-repressor that can't bind any more to the Tet-operator and thus the expression of the autotransporter can be regulated. Although the TetR-system is known to be a tight promoter system there is always a certain background expression with most promoters. If the protein expression of the autotransporter is induced using aTc the autotransporter is transcribed and translated. Due to the bacterial signal peptide the protein is secreted into the bacterial periplasm (the space between the two bacterial membranes of gram-negative bacteria such as E. coli). When present in the bacteria periplasm the autotransporter diffused to the outer membrane and inserts into the membrane. After the integration into the outer membrane the protein cargo is transportet through the beta-barrel of the autotransporter and is pre sented on the bacterial surface. We are sure that this BioBrick will be a valuable contribution to the Parts Registry as it allows future teams to display small protein domains on the surface of gram-negative bacteria, such as E. coli.
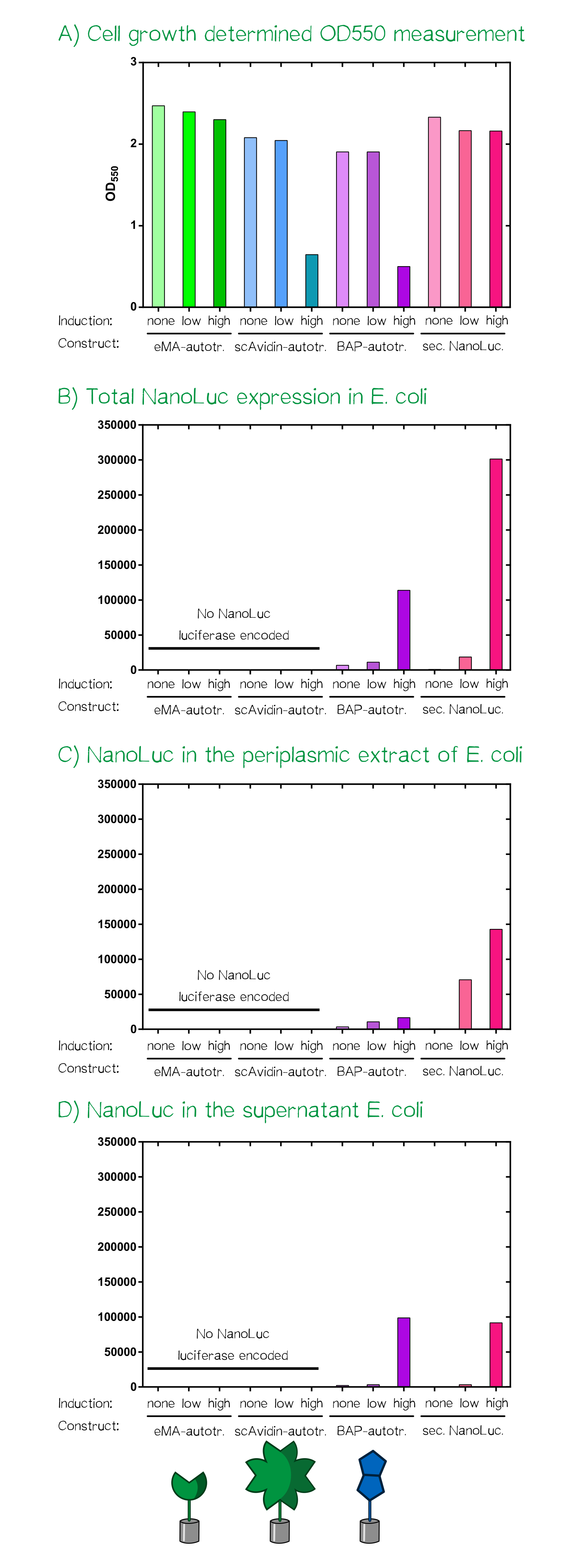
Results: Testing the bacterial autotransporter system
For the testing of our autotransporter constructs we transformed all three autotransporter constructs together with a secreted NanoLuc luciferase, which served as a control, into E. coli JK321[27]. Transformed cells were grown in LB-medium supplemented with chloramphenicol (pSB1C3-vector) at 30°C and 180 RPM. In the morning a stationary over night culture was used to inoculate 50 mL fresh medium with 1 mL of culture. The cultures were grown to OD550 = 0.5 and then the induction of the protein production was started using aTc: "non" = non-induced; "low" = 25 ng/mL aTc; "high" = 200 ng/mL. After induction the cultures were grown for further 5 h at the same concitions and cells were afterwards stored at 4°C until they were used for analysis or bioprinting.
After the cell harvest the optical density (OD550) was determined again as it is known that a high expression of EspP makes the outer membrane of E. coli permeable and this drastically affects cellular viability. For the eMA-autotransporter and the secreted NanoLuc luciferase at high inducer concentration show clearly reduced viability as they did not show any growth after induction at OD550 = 0.5 (see Fig. A). In order to analyze the functionality of the whole design and these new BioBricks we subjected each of the twelve cultures to three different treatments and determined subsequently the amount of NanoLuc luciferase using the manufacturers substrate kit (Thanks to our sponsor Promega ;).
At first the total NanoLuc luciferase content was determined in samples that were just washed once with PBS and contained the whole bacterial cell (see Fig. B), bacterial cells that were incubated with PPA-buffer (periplasmic extraction buffer; 500 mM sucrose, 100 mM Tris/HCl pH 8.0, 1 mM EDTA) that extracts the proteins present in the periplasm (see Fig. C) and finally the culture supernatant of the cultivated bacterial cells (see Fig. D). The total NanoLuc activity is only present in bacteria that are transformed with a plasmid encoding the luciferase and there is a clear dependency of inducer concentration a detected luciferase activity. In the samples where a periplasmic extract was prepared the luciferase activity is highest in the samples of the secreted luciferase as the EspP-fused BAP-NanoLuc is attached to the cells and only proteins from lysed cells are present in the periplasmic extract. In the case of the samples where the supernatant of the cultures was analyzed the luciferase activity is present in both "high" induced samples.
As the luciferase activity only gave an indication for the subcellular localization of the cargo-autotransporter fusion protein, we also performed flow cytometry measurements. For this purpose the same cells were stained with app. 10 µM A3C5-Fab that was coupled to the fluorescent dye Dy634-NHS and 100 µM of the chemical compound biotin-atto488. Cells were incubated for 2 h at 4°C, washed twice with PBS and analyzed using a FACSAria II flow cytometer (BD bioscience, voltages for fluorescence channels were all at 750 V).
References
- ↑ Chen, I., Howarth, M., Lin, W., & Ting, A. Y. (2005). Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nature methods, 2(2), 99-104.
- ↑ Kozak, M. (1989). The scanning model for translation: an update. The Journal of cell biology, 108(2), 229-241.
- ↑ Kozak, M. (1987). An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic acids research, 15(20), 8125-8148.
- ↑ Petersen, T. N., Brunak, S., von Heijne, G., & Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature methods, 8(10), 785-786.
- ↑ Sonnhammer, E. L., Von Heijne, G., & Krogh, A. (1998, July). A hidden Markov model for predicting transmembrane helices in protein sequences. In Ismb (Vol. 6, pp. 175-182).
- ↑ Alexander, H., Harpprecht, J., Podzuweit, H. G., Rautenberg, P., & Müller-Ruchholtz, W. (1994). Human monoclonal antibodies recognize early and late viral proteins of human cytomegalovirus. Human Antibodies, 5(1-2), 81-90.
- ↑ Kredel, S., Oswald, F., Nienhaus, K., Deuschle, K., Röcker, C., Wolff, M., ... & Wiedenmann, J. (2009). mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PloS one, 4(2), e4391.
- ↑ Bajar, B. T., Wang, E. S., Lam, A. J., Kim, B. B., Jacobs, C. L., Howe, E. S., ... & Chu, J. (2016). Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Scientific reports, 6.
- ↑ Schmidt, T. G., & Skerra, A. (2007). The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nature protocols, 2(6), 1528-1535.
- ↑ Case, J. F. (2004). Flight studies on photic communication by the firefly Photinus pyralis. Integrative and comparative biology, 44(3), 250-258.
- ↑ Zhang, L., Song, G., Xu, T., Wu, Q. P., Shao, X. X., Liu, Y. L., ... & Guo, Z. Y. (2013). A novel ultrasensitive bioluminescent receptor-binding assay of INSL3 through chemical conjugation with nanoluciferase. Biochimie, 95(12), 2454-2459.
- ↑ Gould, S. J., & Subramani, S. (1988). Firefly luciferase as a tool in molecular and cell biology. Analytical biochemistry, 175(1), 5-13.
- ↑ Hall, M. P., Unch, J., Binkowski, B. F., Valley, M. P., Butler, B. L., Wood, M. G., ... & Robers, M. B. (2012). Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS chemical biology, 7(11), 1848-1857.
- ↑ Nanbo, A., Sugden, A., & Sugden, B. (2007). The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. The EMBO journal, 26(19), 4252-4262.
- ↑ Boussif, O., Lezoualc'h, F., Zanta, M. A., Mergny, M. D., Scherman, D., Demeneix, B., & Behr, J. P. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine.Proceedings of the National Academy of Sciences, 92(16), 7297-7301.
- ↑ Molina, D. K., & DiMaio, V. J. (2012). Normal organ weights in men: part II—the brain, lungs, liver, spleen, and kidneys. The American journal of forensic medicine and pathology, 33(4), 368-372.
- ↑ Wilson, Z. E., Rostami‐Hodjegan, A., Burn, J. L., Tooley, A., Boyle, J., Ellis, S. W., & Tucker, G. T. (2003). Inter‐individual variability in levels of human microsomal protein and hepatocellularity per gram of liver. British journal of clinical pharmacology, 56(4), 433-440.
- ↑ https://www.thermofisher.com/de/de/home/references/protocols/proteins-expression-isolation-and-analysis/protein-expression-protocol/flp-in-system-for-generating-constitutive-expression-cell-lines.html#prot4
- ↑ Blochlinger, K. A. R. E. N., & Diggelmann, H. E. I. D. I. (1984). Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Molecular and cellular biology, 4(12), 2929-2931.
- ↑ Livak, Kenneth J., and Thomas D. Schmittgen. "Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method." methods 25.4 (2001): 402-408.
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an Anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of molecular biology, 400(4), 783-802.
- ↑ Ghrayeb, J., Kimura, H., Takahara, M., Hsiung, H., Masui, Y., & Inouye, M. (1984). Secretion cloning vectors in Escherichia coli. The EMBO journal, 3(10), 2437.
- ↑ Costa, J., Grabenhorst, E., Nimtz, M., & Conradt, H. S. (1997). Stable expression of the Golgi form and secretory variants of human fucosyltransferase III from BHK-21 cells Purification and characterization of an engineered truncated form from the culture medium. Journal of Biological Chemistry, 272(17), 11613-11621.
- ↑ Barnard, T. J., Dautin, N., Lukacik, P., Bernstein, H. D., & Buchanan, S. K. (2007). Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nature structural & molecular biology, 14(12), 1214-1220.
- ↑ Skillman, K. M., Barnard, T. J., Peterson, J. H., Ghirlando, R., & Bernstein, H. D. (2005). Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Molecular microbiology, 58(4), 945-958.
- ↑ Jose, J., Krämer, J., Klauser, T., Pohlner, J., & Meyer, T. F. (1996). Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga β autotransporter pathway. Gene, 178(1), 107-110.