(→Medical Application) |
|||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
=Medical Application= | =Medical Application= | ||
| + | ==Therapeutic Application ≠ usage of fancy state-of-the-art methods BUT reliable simplistic methods== | ||
| + | |||
| + | In Biomedical Research, only transparent and simplistic therapeutic approaches, which can be evaluated in all stages of clinical trials, will end up in the patient. | ||
| + | |||
| + | Nowadays, one of the holy grail in Synthetic Biology is a) to understand the complex regulation mechanism on (co)-transcriptional and (co-)translational networks on every imaginable level and b) to manipulate and rewire those networks. | ||
| + | |||
| + | Great efforts, especially in Computational Biology and Systems Biology, have been achieved to understand the complex homeostasis-network of life. Although many principles of regulation have been understand, only relative small networks can be modelled and simulated computationally; the outcome of a “healthy” network are well understood in this case. | ||
| + | |||
| + | BUT pathological networks, where a defective network node cause a shift in the cell/organ/organisms’ homeostasis and thus result in “disease” are poorly understood, since there are still too many unknown network nodes and/or the dynamic and impact of a node is not understood | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
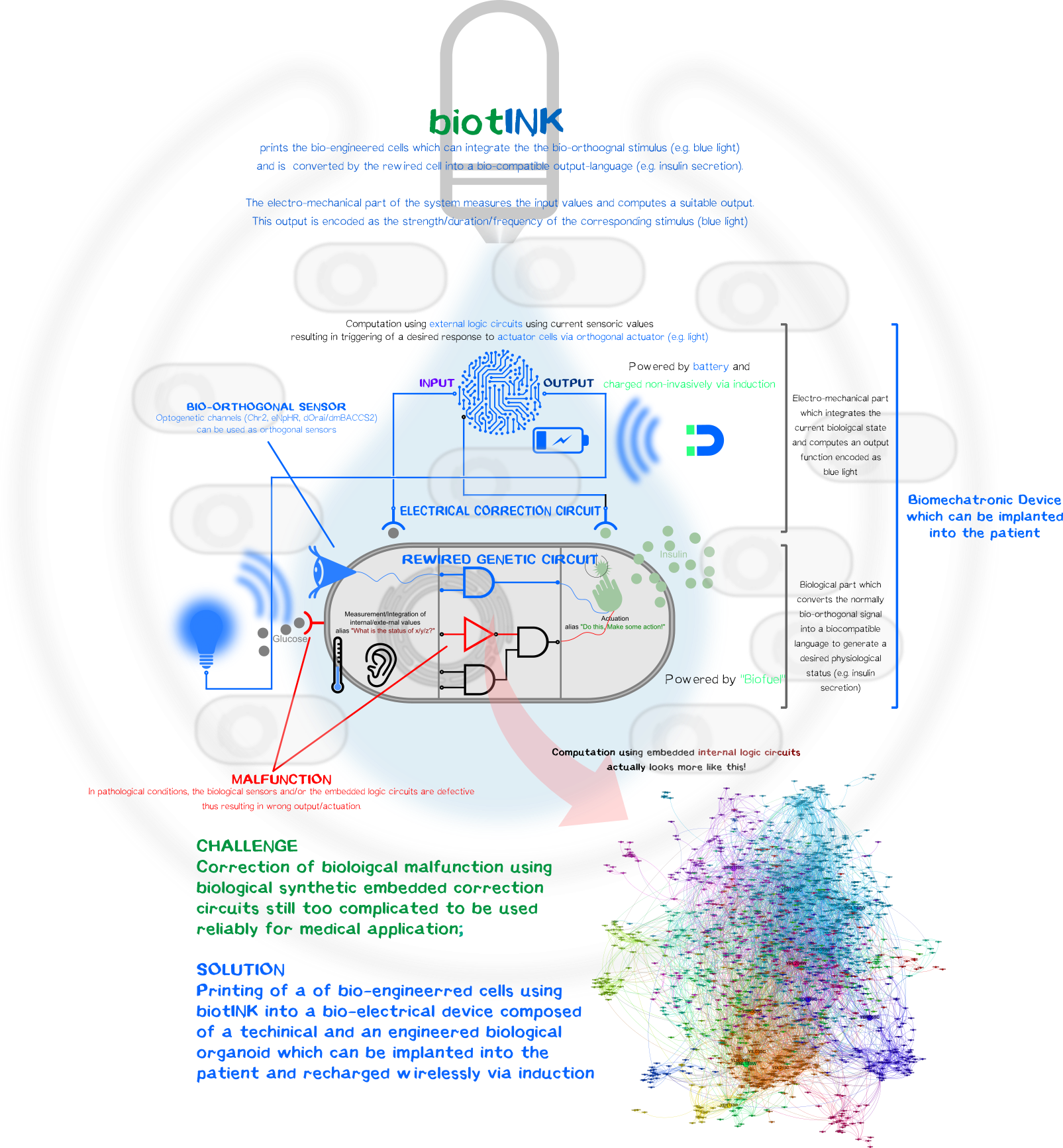
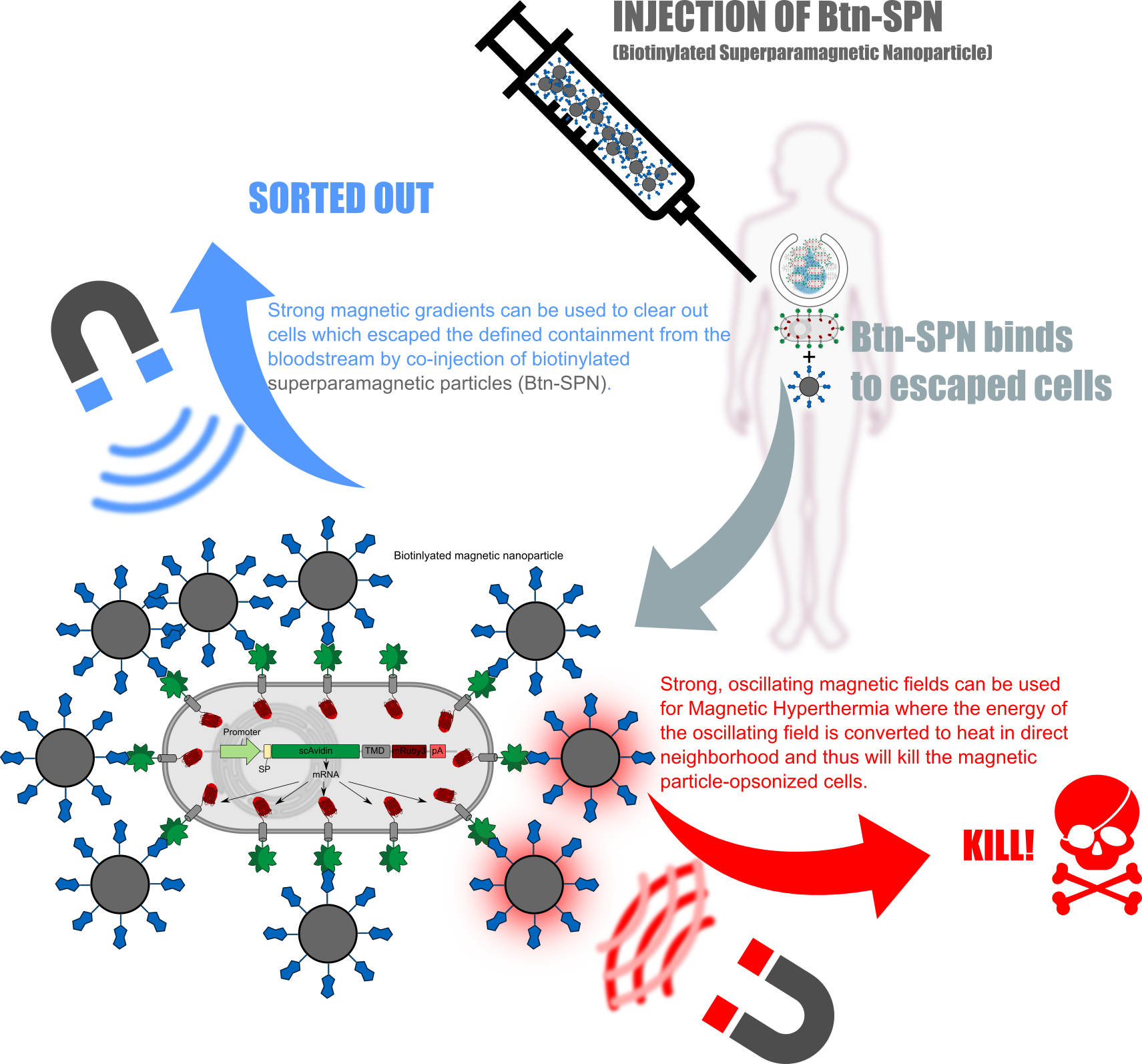
[[File:T--LMU-TUM_Munich--MedApp_Fig1.png|thumb|center|850px| | [[File:T--LMU-TUM_Munich--MedApp_Fig1.png|thumb|center|850px| | ||
Revision as of 19:07, 19 October 2016
Medical Application
Therapeutic Application ≠ usage of fancy state-of-the-art methods BUT reliable simplistic methods
In Biomedical Research, only transparent and simplistic therapeutic approaches, which can be evaluated in all stages of clinical trials, will end up in the patient.
Nowadays, one of the holy grail in Synthetic Biology is a) to understand the complex regulation mechanism on (co)-transcriptional and (co-)translational networks on every imaginable level and b) to manipulate and rewire those networks.
Great efforts, especially in Computational Biology and Systems Biology, have been achieved to understand the complex homeostasis-network of life. Although many principles of regulation have been understand, only relative small networks can be modelled and simulated computationally; the outcome of a “healthy” network are well understood in this case.
BUT pathological networks, where a defective network node cause a shift in the cell/organ/organisms’ homeostasis and thus result in “disease” are poorly understood, since there are still too many unknown network nodes and/or the dynamic and impact of a node is not understood
Seitenverantwortliche/r:Christoph
Literaturreferenz
Literaturreferenz[1]
Bei Google Scholar bitte das APA-Ziteirformat verwenden.
Textformatierung
kursiv
fett
Strich
Links
Wikiinterner Link Team:LMU-TUM_Munich/Materials (As described in the Materials section)
Wikiexterner Link Visit W3Schools
Visit W3Schools
Bilder
Introduction
Design
Experiments
Proof of concept
Demonstrate
Discussion
References
- ↑ Schmidt, T. G., & Skerra, A. (2007). The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nature protocols, 2(6), 1528-1535.