(→BioBrick-compatible vectors for mammalian cells: transient tranfection & stable integration) |
|||
| Line 57: | Line 57: | ||
=Subcellular protein localization using fluorescence microscopy= | =Subcellular protein localization using fluorescence microscopy= | ||
| + | [[File:Muc16 Receptor_Microscopy_001.png |thumb|right|850px]] | ||
| + | |||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | |||
=Quantification of functionalized membrane proteins using flow cytometry= | =Quantification of functionalized membrane proteins using flow cytometry= | ||
| + | [[File:Muc16 Receptor_FACS_001.png |thumb|right|850px]] | ||
| + | |||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | |||
=Immunochemical detecton of functionalized membrane protein using Western Blot= | =Immunochemical detecton of functionalized membrane protein using Western Blot= | ||
| + | [[File:Muc16 Receptor_WB_001.png |thumb|right|850px]] | ||
| + | |||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | |||
{| | {| | ||
|+ Properties of membrane proteins | |+ Properties of membrane proteins | ||
| Line 88: | Line 121: | ||
=High resolution imaging using scanning electron microscopy (SEM)= | =High resolution imaging using scanning electron microscopy (SEM)= | ||
| + | [[File:Muc16 Receptor_SEM_001.png |thumb|right|850px]] | ||
| + | |||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
| + | <br><br><br><br> | ||
=References= | =References= | ||
Revision as of 22:29, 15 October 2016
Overview
Protein targeting
Essential to a protein's function is not only its activity or binding properties, but also its presence at the right time, at the right place. Just like in real estate, the motto here is 'location, location, location'. One of the most ubiquitous elements for protein targeting is the signal peptide, which enables the translocation of proteins to the ER, from where their journey may continue to the place of function. Proteins containing a signal peptide include transmembrane proteins, secreted proteins, proteins of the ER itself, proteins of the Golgi apparatus and several more. For our project, signal peptides constitute a crucial part for many constructs, being used for targeting of transmembrane proteins to the cell surface, targeting proteins to the ER and the secretion of proteins.
Signal peptides
The key for the secretory pathway: signal peptides
At the beginning of the 1970s, later Nobel Prize laureate Guenther Blobel made the discovery that some in vitro translated proteins would turn out slightly longer than their counterparts found in cells. This observation led to the discovery of the signal peptide, a short N-terminal protein sequence that is required for the targeting of proteins into the endoplasmic reticulum (ER) where it is cleaved off afterwards. This mainly hydrophobic sequence, being both required and sufficient for membrane proteins to be translocated to their place of action, allows targeting of proteins to the ER and to their specific location in the membrane - in the case of receptors to the cell surface. The signal peptide is a short peptide (up to 30 amino acids) that is recognized by the so-called signal recognition particle upon translation at the rough ER, which mediates the co-translational translocation of the protein’s peptide chains into the ER via a translocon called the Sec-complex. During this project, three different peptides are being tested. The EGFR signal peptide was taken from the iGEM parts registry (BBa_K157001) and combined with the CMV-promoter via the RFC10 cloning standard, while CMV-BM40-SP and CMV-IgKappa constructs were designed manually and synthesized, and combined with a BioBrick containing the CMV promoter sequence via restriction cloning. As shown in figure X, the EGFR signal peptide taken from the parts registry is constructed in a way that the signal peptide ORF immediately follows the cloning scar after the CMV promoter, thus resulting in a very short 5’ untranslated region (UTR) of the transcribed mRNA. The CMV-signal peptide constructs created for the project on the other hand, which contain the BM40 and IgKappa signal peptides, were designed to have a considerably longer 5’ UTR, additionally allowing them to contain a full Kozak consensus sequence. The Kozak sequence is recognized by the ribosome as a translational start site; this element missing or deviating from the consensus sequence may considerably decrease translation efficiency. For the EGFR signal peptide construct, a Kozak sequence is not present, as it would have to preceed the start codon ATG - a position which is occupied by the RFC10 cloning scar. Since both the distance from the promoter to the open reading frame as well as the Kozak consensus sequence are considered crucial parameters for expression levels, the BM40 and IgKappa constructs were designed to potentially increase expression levels of the receptor.
For testing the functionality of the signal peptide, two different approaches are taken: three non-receptor constructs were cloned (not shown), only containing the CMV promoter, one out of the three signal peptides, a nanoluciferase, Strep- tag II as well as a the hGH polyadenylation sequence. By measuring the secretion of the fusion protein into the medium via a luciferase assay, one can quantify the expression- and secretion levels of the nanoluciferase-fusion protein and thus the functionality of the signal peptides. A second approach for determining signal peptide functionality utilizes the bioinformatic tool SignalP, which can assign theoretical scores for the functionality of signal peptides as well as their cleavage site.
Bioinformatic prediction of signal peptide properties using the SignalP server
As the choice of signal peptide was not obvious from the beginning, the three options (IgKappa, BM40, EGFR) were tested via bioinformatic tools as well as an assay of a secretion fusion protein containing the respective signal peptides. Concerning the theoretical prediction of signal peptide efficiency, SignalP is able to determine whether a sequence is likely to function as a signal peptide, as well as predict the probable cleavage site of the signal peptide after its translocation into the ER. The results of SignalP prediction are shown in figure X. For all constructs, signal peptide functionality is predicted for the N-terminal region, as well as a potential cleavage site. The SignalP 4.1 server was used for theoretical prediction of signal peptide properties (http://www.cbs.dtu.dk/services/SignalP). The complete translated amino acid sequences of the respective receptor constructs were used as input. The algorithm for eukaryotes with default D-cutoff value was chosen.
For all three signal peptides, the high S-score indicates general signal peptide functionality. For the IgKappa signal peptide, a cleavage site between amino acid 20 and 21 within the signal peptide is predicted. For the BM40 signal peptide, a cleavage site between amino acid 17 and 18 is predicted, and for the EGFR signal peptide, a cleavage site between amino acid 24 and 25 is predicted.
Results: Design of signal peptides and characterization

For the quantification of signal peptide functionality via the first approach, constructs only containing the CMV promoter, one of three signal peptides - the EGFR signal peptide, the Igκ signal peptide or the BM40 signal peptide, a nanoluciferase, a Strep-tag II, and the hGH polyadenylation signal sequence were used. Lacking a transmembrane domain but containing a signal peptide, these constructs are being translocated into the ER and - not containing any further targeting signal - then secreted into the medium. Using the luciferase assay, one can quantifiy the amount of luminescence - and thus, proportionally, the amount of secreted nanoluciferase - by measuring the conversion of luciferin into visible light and integrating it over a certain timespan.
The result of the assay are shown in figure X. The BM40 signal peptide shows the highest integrated fluorescence signal at all time points, indicating the highest amount of secretion of the nanoluciferase construct and thus highest signal peptide functionality.
Transmembrane domains
BioBrick-compatible vectors for mammalian cells: transient tranfection & stable integration
- Abbildung pDSG/pcDNA5 vektoren
- Abbilgung mit stabiler Integration, Zeocin-Test, genomische PCR
Subcellular protein localization using fluorescence microscopy
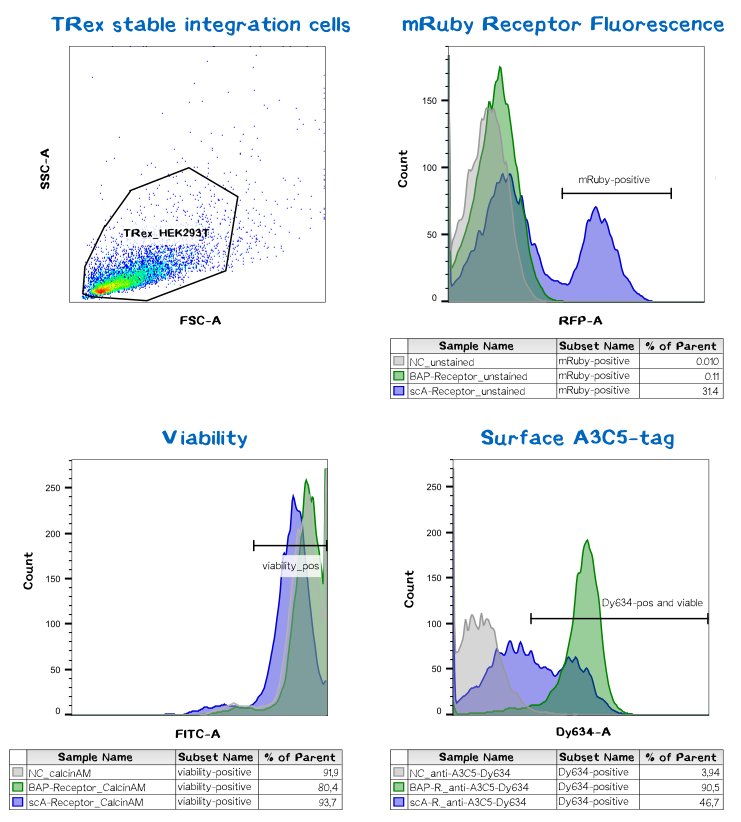
Quantification of functionalized membrane proteins using flow cytometry
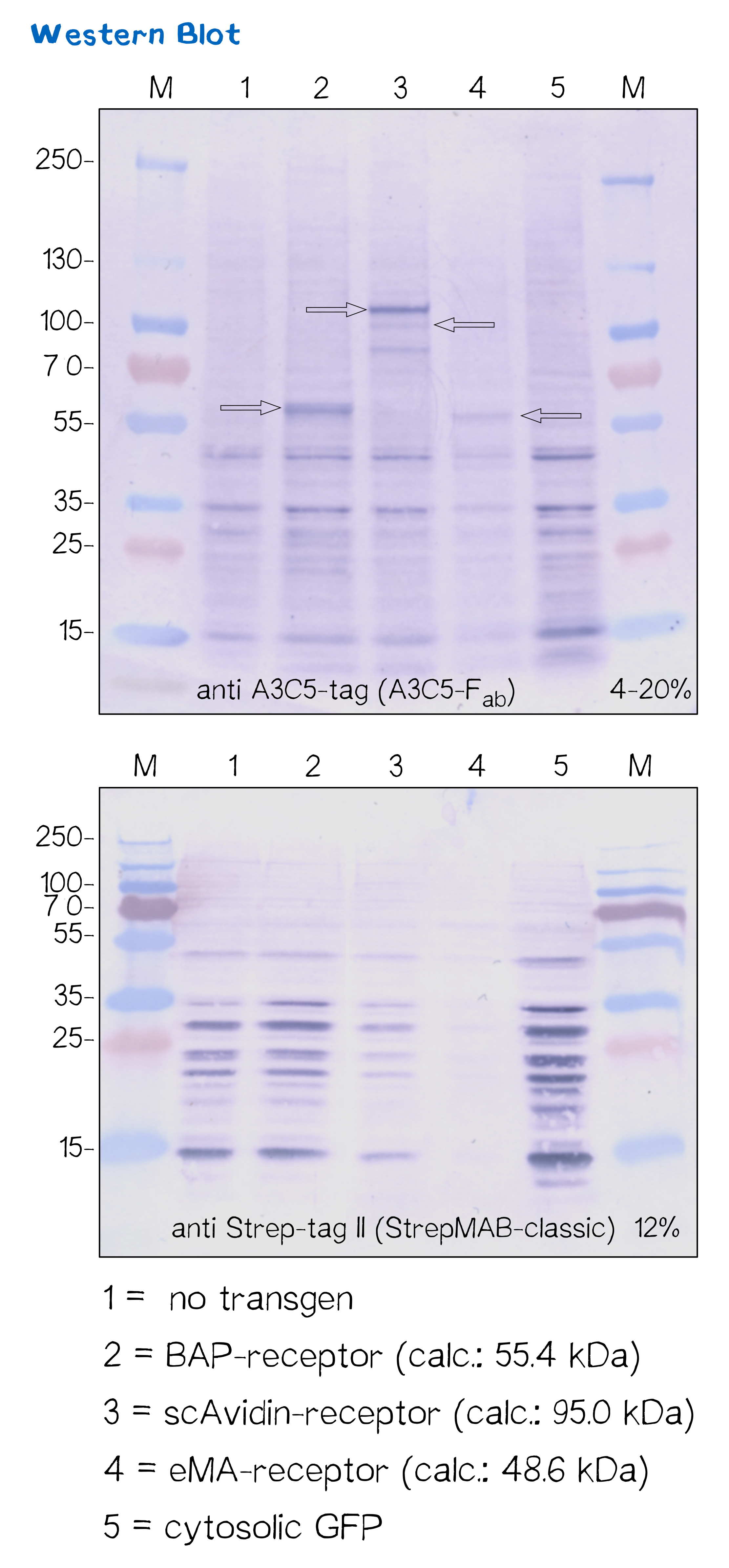
Immunochemical detecton of functionalized membrane protein using Western Blot
| Receptor | BioBrick | Amino acids | Moelcular mass [Da] |
| BAP-Receptor | [http://parts.igem.org/Part:BBa_K2170000 BBa_K2170000] | 507 | 55 440 |
| eMA-Receptor | [http://parts.igem.org/Part:BBa_K2170001 BBa_K2170001] | 448 | 48 618 |
| scAvidin-Receptor | [http://parts.igem.org/Part:BBa_K2170002 BBa_K2170002] | 877 | 95 028 |
Quantification of mRNA expression levels by RT-q-PCR
High resolution imaging using scanning electron microscopy (SEM)
References