Design
Design as a whole
The conception of synthetic biology has been universally employed in detection and quantitative measurement of the levels of extracellular and intracellular molecules. In order to make people capable of knowing their health conditions so that they can figure out what is going wrong at early stage, our goal has always been to create an in vitro yeast disease biosensor which allows people to:
- Use the biosensor anytime and anywhere they want
- Obtain their results as fast as possible
- Monitor their disease biomarkers at a range as wide as possible
- Get the device as cheap as possible
To achieve our goal, we managed to construct the biosensor for diagnosis of two diseases, diabetes and phaeochromocytoma, using urine sugar and epinephrine as their biomarkers. Three major types of elements to genetically modify the yeast Saccharomyces cerevisiae.: The first type of elements is promoter, they are used to control the expression of other elements in yeast; the second type of elements is reporter protein, Nano-lantern(cAMP-1.6), which allows detection of cellular response to extracellular ligands within 10 minutes; the third type of elements is G protein coupled receptor, ADRB2, which allows detection of the disease biomarker of phaeochromocytoma, excreted epinephrine.
The overall constructing process can be figured as bellow:
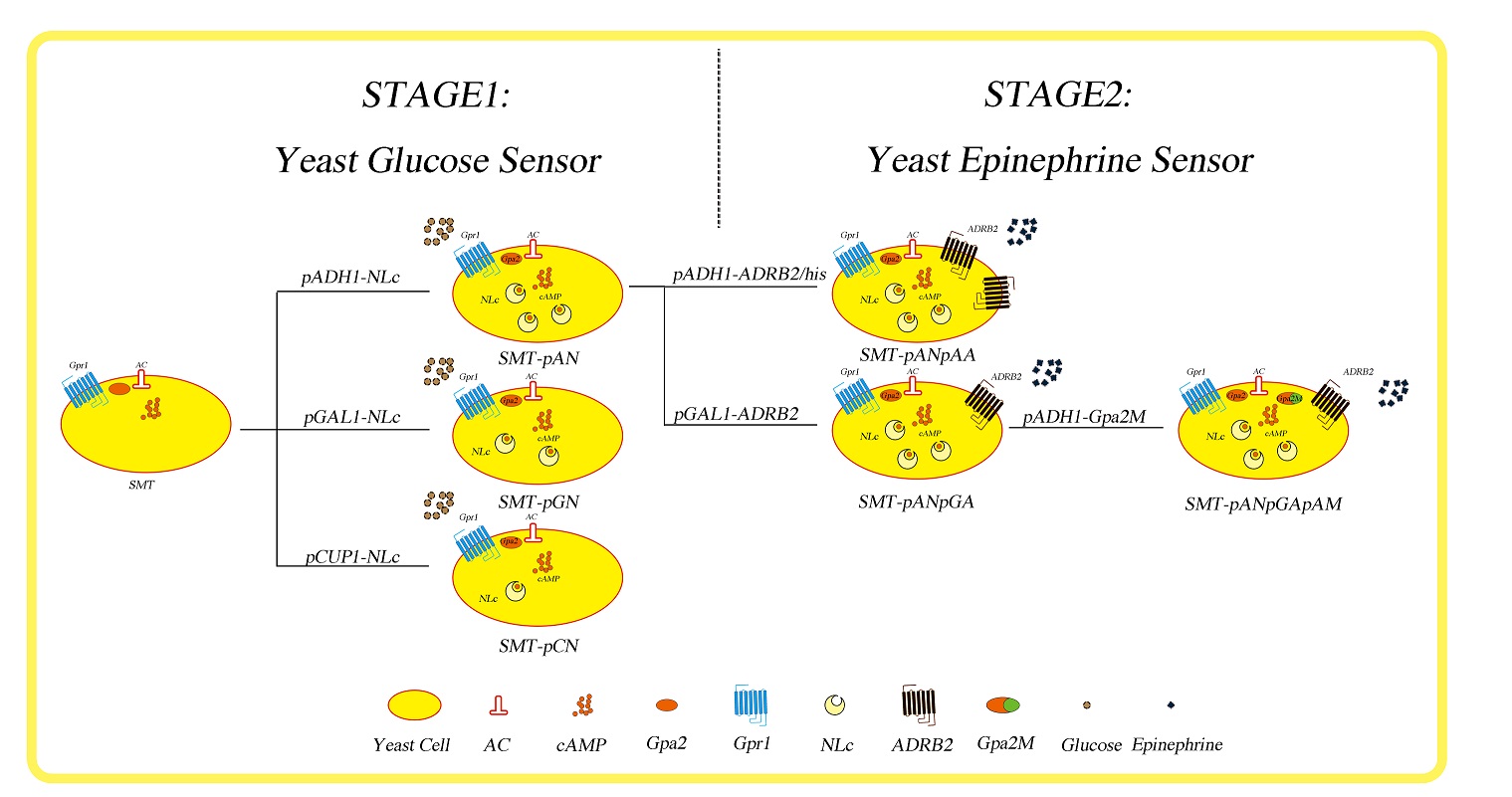
Fig.1 The construct process of real-time yeast disease biosensor. SMT: our chassis yeast cell; AC: adenylyl cyclase; cAMP: cyclic adenosine monophosphate; Gpa2: Saccharomyces cerevisiae S288c guanine nucleotide-binding protein subunit alpha; Gpr1: plasma membrane G protein coupled receptor (GPCR) for glucose sensing; NLc: Nano-lantern (cAMP 1.6); ADRB2: human adrenergic receptor beta 2; Gpa2M: S. cerevisiae Gpa2 with 5 amino acids from human G(s) alpha-subunit.
As shown in figure 1, we named our chassis yeast cell ‘SMT’, which stands for ‘smart’. At the first stage, where our goal is to create a real-time yeast glucose sensor, we utilized the endogenous yeast glucose sensing mechanism, which starts with the binding of glucose to Gpr1 receptor and ends with the altered gene expression. The genotype of SMT is MATα his3Δ leu2Δ lys2Δ ura3Δ;pho8Δ60,pho13Δ,trpΔ, and thus allows us to make genomic insertions and selection at HIS3, LEU2, LYS2, URA3 and TRP1 locus. So we cloned a cAMP sensing protein, Nano-lantern (cAMP 1.6) downstream three kinds of yeast endogenous promoters, pADH1, pGAL1 and pCUP1 into pBluescript with homologous sequence of TRP1, then we made genomic insertion at TRP1 locus in SMT cells yielding strains SMT-pAN, SMT-pGN and SMT-pCN, respectively. Then at stage 2, creating yeast epinephrine sensor, we managed to clone human adrenergic receptor beta-2 (ADRB2) downstream ADH1 and GAL1 promoter and inserted these cassettes at HIS3 locus of SMT-pAN yielding strains SMT-pANpAA and SMT-pANpGA. Finally, we tried to used modified yeast G alpha subunit, Gpa2, to increase the efficiency of functional coupling between heterologously expressed ADRB2 and yeast endogenous signaling pathway, but we didn’t go any further than making the right insertion construct pADH1-Gpa2M as time was limited.
Why yeast?
Cell-based biosensing, in general, and yeast-based biosensing (YBB) in particular have many advantages for certain diagnostic and analytical applications. High-specificity biosensing can be carried out in a micron-scale unit (the cell) without power requirements or the need for a highly trained operator. This can be contrasted with a Western blot or a gas chromatograph mass-spectrometer (GCMS), which are orders-of-magnitude larger, and require power and operators[1]. Moreover, the existing testing methods only apply to a quite limit range of non-molecular or molecular indices, like blood pressure, blood sugar and HCG, and if one wish to detect a wider range of markers, the prices will soar up because of the high expenditure of ELISA based chips.
First, as a eukaryotic cell, yeast Saccharomyces cerevisiae is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like Escherichia coli as the model bacterium. Being eukaryotic cell makes S. cerevisiae capable of serving as a chassis for genetic engineering involves mammalian sensing elements, such as G-protein coupled receptors and nuclear receptors[1, 2]. Second, it can be made in ‘active dry’ form cheaply and stored for long periods of time, thus the shelf-life of yeast biosensor makes it a potential in vitro diagnostic biosensor[2].
Increase the versatility and composability of the biosensor
There are millions of way that one can choose when coming across the topic of constructing a whole cell disease biosensor. Yeast is an excellent eukaryotic chassis which can be used to assemble diversity of sensor molecules, and so as to increase the composability, we chose the G-protein couple receptor as our sensing element because of its highly patterned structure and incredibly wide range of ligands.
The G-protein-coupled receptors (GPCRs) constitute a large superfamily of transmembrane proteins that mediate cellular responses to diverse extracellular stimuli, including light, neurotransmitters, hormones and odorants. Through selective ligand binding, the GPCRs discriminate between these multiple external signals. They share the common structure of seven transmembrane-spanning domains[3]. The GPCRs amplify and transduce the information inherent in ligand binding to the cell interior via interaction with intracellular heterotrimeric G-proteins. As a result, the G-protein complex undergoes guanine-nucleotide exchange and subunit dissociation in response to the agonist. After activation by agonist-bound GPCR, the GTP-bound alpha subunit and the beta-gamma complex are free to modulate the activity of a variety of effector proteins, including adenylyl cyclase, phospholipase C, G-protein-gated Ca2+ and K+ channels, and membrane-proximal elements of mitogen-activated protein kinase (MAP kinase) signal-transduction pathways[4]. A typical signaling pathway of GPCR can be picturized as follows:
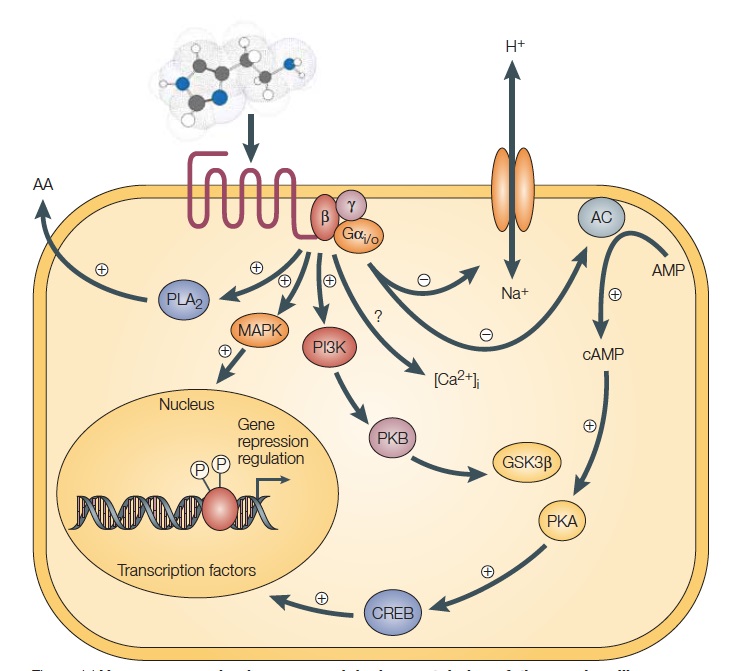
Fig.2 H3-receptor signaling pathways. Source: Leurs, R. et al. The histamine H3 receptor: from gene cloning to H3 receptor drugs. Nature Reviews Drug Discovery 4, 107-120 (2005).
Our current knowledge about GPCRs and their natural ligands is the combined work of several decades of research. However, we have only come half the distance, or even less. Cloning of the human genome has revealed about 1000 GPCRs—400 for neurotransmitters and hormones (transmitters, as a general term) and about 600 presumed odorant and gustatory receptors (sensory receptors). [5]. In our project, we utilized 2 kinds of GPCRs, namely Gpr1 from Saccharomyces cerevisiae for glucose sensing and ADRB2 from Homo sapiens for catecholamine sensing, they are picturized as follows [6, 7]:
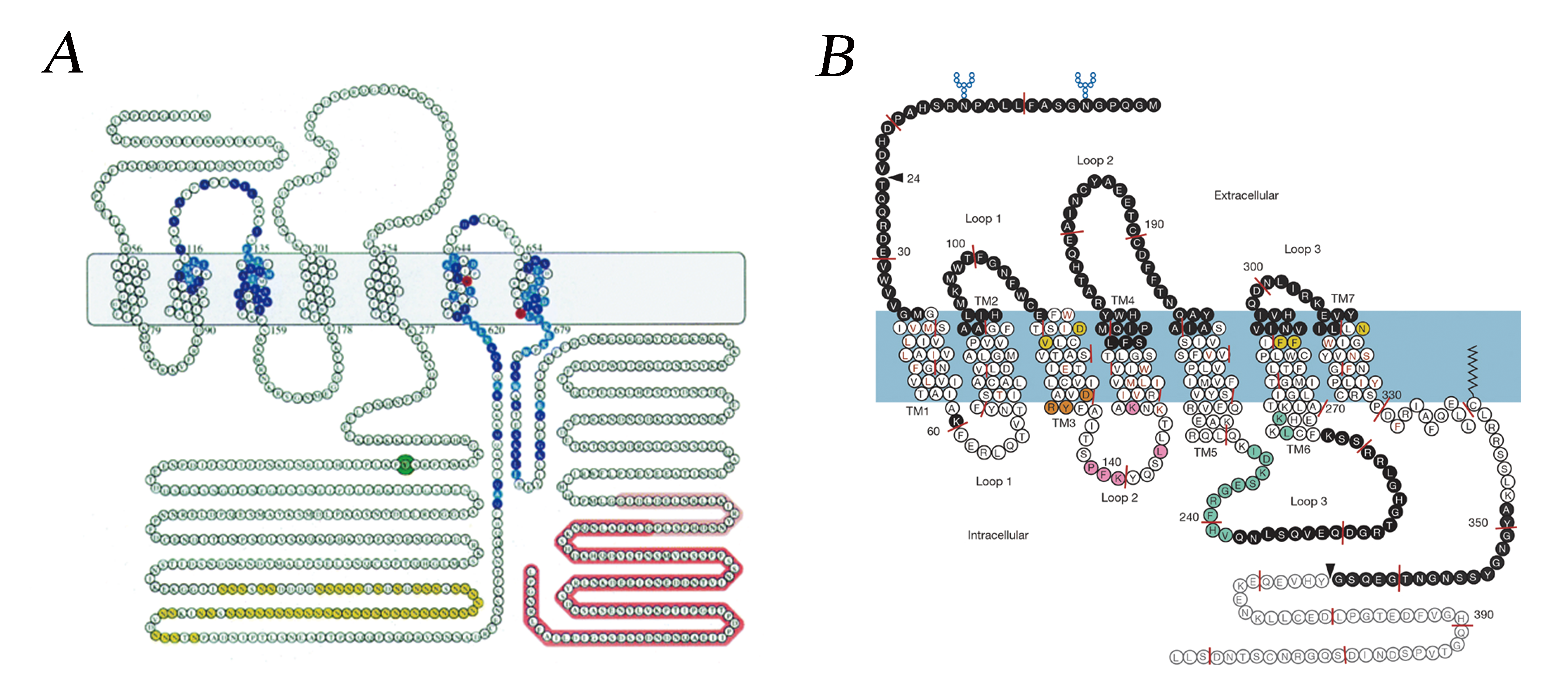
Fig.3 Gpr1 from Saccharomyces cerevisiae (A) and ADRB2 from Homo sapiens (B).
Increase the response speed of whole cell biosensor
The majority of yeast-based biosensors use a reporter gene under the control of an inducible promoter. The promoters are activated in the presence of a ligand of interest, either directly by the ligand/receptor dimer or via a signaling cascade. The most common reporting systems for yeast biosensors are fluorescence, luminescence, colorimetry, electrical methods and growth rate[1]. However, nearly all of the existing yeast biosensors are transcription-based, which means they need a long time to yield the detectable sequence, even though previous researchers have enhanced the signal coupling ability of heterologous expressed GPCRs, the response and culture time make it hard to be operated by ordinary individuals and served as a domestic testing chip[5, 8, 9]. Though several transcripton-independent sensors have been created, the indices that can be detected still remain limited[1, 10].
As we were going to use the GPCR signaling pathway as our sensing mechanism, we looked for molecules downstream the GPCR from G-proteins to transcription factors which can be changed rapidly after binding with the ligands. cAMP serves as a second messenger in cellular signaling, and its intracellular level can be soared very rapidly (within 10 min) with the catalysis of adenylyl cyclase, which is activated by G protein alpha subunit when the ligands bind corresponding GPCR. So we sought to find a way to indicate the intracellular cAMP level to reveal the extracellular ligand level.
An excellent cAMP indicator has been developed by professor Nagai, it is a fusion protein named Nano-lantern(cAMP-1.6), it requires no excitation light and can be observed under optical microscope[11]. Nano-lantern(cAMP-1.6) contains a portion of enhance YFP with 10 amino acids deleted at C terminus, denoted as Venus△C10, a portion of mutated Renilla luciferase with 3 amino acids deleted at N terminus, Rluc8, and a cAMP binding domain of EPAC1 (Exchange Protein Directly Activated by cAMP) flanked by two separate parts of Rluc8. Upon binding with the cAMP molecule, the catalytic activity of the split luciferase will increase as the separate parts are brought together because of the conformational change in EPAC, thus results in the change in luminescence via the BRET effect. The construct of the protein can be picturized as below:
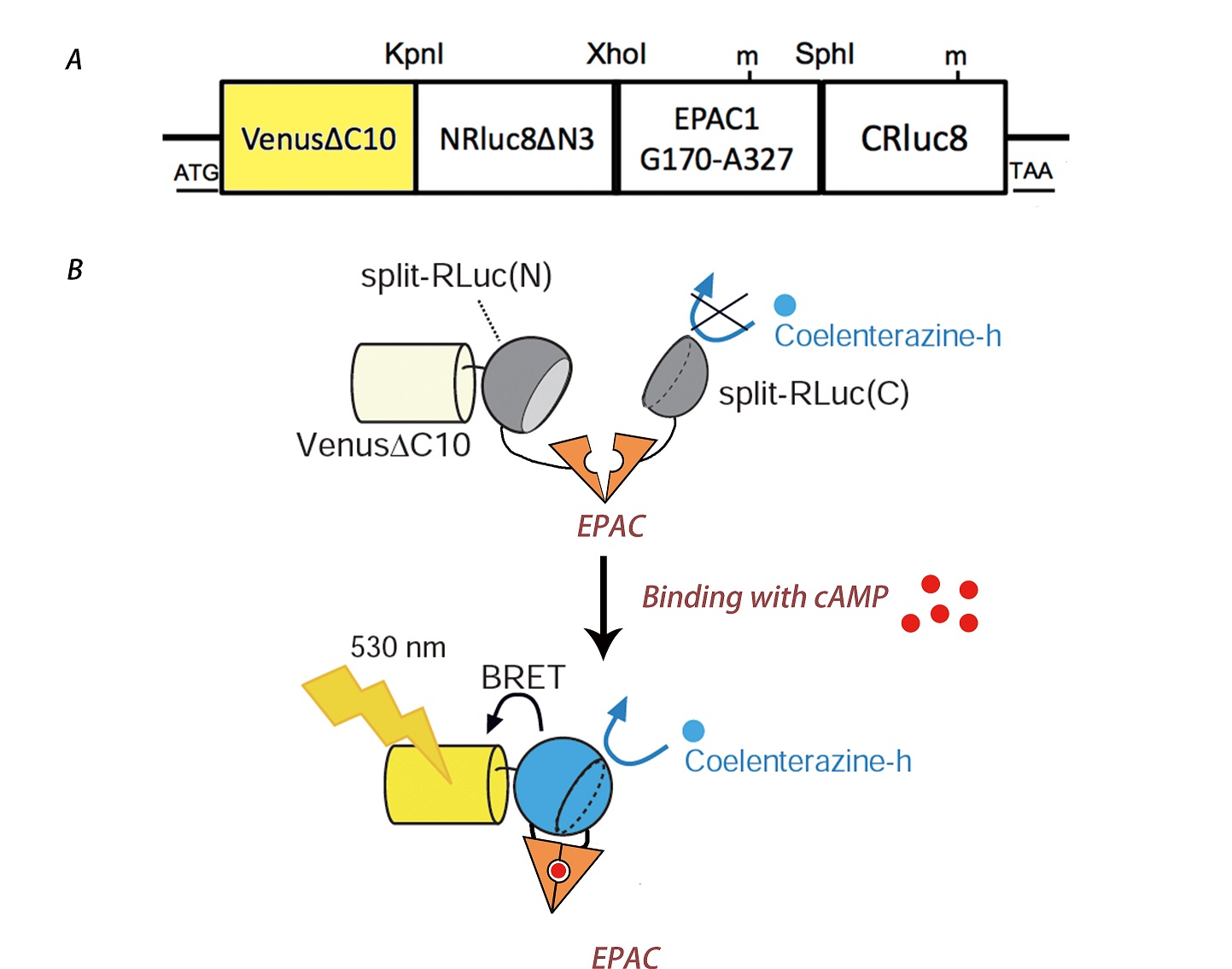
Fig.4 The construct detail of Nano-lantern(cAMP-1.6) (A) and its mechanism (B).
Increase the shelf life of yeast product
Freeze-drying is a two-step dehydration process used for the long-term preservation of perishable materials, including living cells. This method has been relatively well characterized and successfully applied in some bacteria-based portable biosensors, but few information is available for similar approaches in yeast. A research group from Czech Republic has employed this technology for their ER/AR yeast biosensors[2]. Our project characterized the applicability of freeze-drying methods in our real-time yeast biosensors and proposed plans for future optimization.
Reference
[1] Adeniran, A., M. Sherer, and K.E. Tyo, Yeast-based biosensors: design and applications. FEMS Yeast Res, 2014.
[2] Jarque, S., M. Bittner, and K. Hilscherova, Freeze-drying as suitable method to achieve ready-to-use yeast biosensors for androgenic and estrogenic compounds. Chemosphere, 2016. 148: p. 204-210.
[3] O'Malley, M.A., et al., Progress toward heterologous expression of active G-protein-coupled receptors in Saccharomyces cerevisiae: Linking cellular stress response with translocation and trafficking. Protein Science, 2009. 18(11): p. 2356-2370.
[4] Pausch, M.H., G-protein-coupled receptors in Saccharomyces cerevisiae: high-throughput screening assays for drug discovery. Trends in Biotechnology, 1997. 15(12): p. 487-494.
[5] Mukherjee, K., S. Bhattacharyya, and P. Peralta-Yahya, GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. Acs Synthetic Biology, 2015. 4(12): p. 1261-1269.
[6] Kraakman, L., et al., A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Molecular Microbiology, 1999. 32(5): p. 1002-1012.
[7] Rasmussen, S.G., et al., Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature, 2007. 450(7168): p. 383-7.
[8] Fukuda, N., et al., Amplification of agonist stimulation of human G-protein-coupled receptor signaling in yeast. Analytical Biochemistry, 2011. 417(2): p. 182-187.
[9] Ishii, J., et al., Microbial fluorescence sensing for human neurotensin receptor type 1 using G alpha-engineered yeast cells. Analytical Biochemistry, 2014. 446: p. 37-43.
[10] Zhang, Y.F., et al., pHlash: A New Genetically Encoded and Ratiometric Luminescence Sensor of Intracellular pH. Plos One, 2012. 7(8).
[11] Saito, K., et al., Luminescent proteins for high-speed single-cell and whole-body imaging. Nature Communications, 2012. 3.




