In the notebook, there are some simplifications we made in sample names:
| Orignal | Simplified |
| ADH1 promoter( pADH1) | ADH |
| GAL1 promoter (pGAL1) | GAL |
| CUP1 promoter (pCUP1) | CU |
| Nano-lantern cAMP 1.6 | NLc |
| human adrenergic receptor beta 2 (ADRB2) | hAR |
| yeast nucleotide binding alpha subunit (Gpa2) with 5 amino acids modification | Gpa2M |
| Yeast TRP section marker | TRP |
| Yeast HIS section marker | HIS |
| Yeast LEU section marker | LEU |
| 6 x His-tag | his |
20160624
- Designed primers for amplifying the ADH1 promoter from yeast genome .
- Our oder (Nano-lantern cAMP 1.6) from addgene arrived!
20160625
- pick up colonies of pcDNA3-Nanolantern.
- Plasmid extraction of RS416、426、424、LD-BS-ADH1-COS1( plasmid backbones)
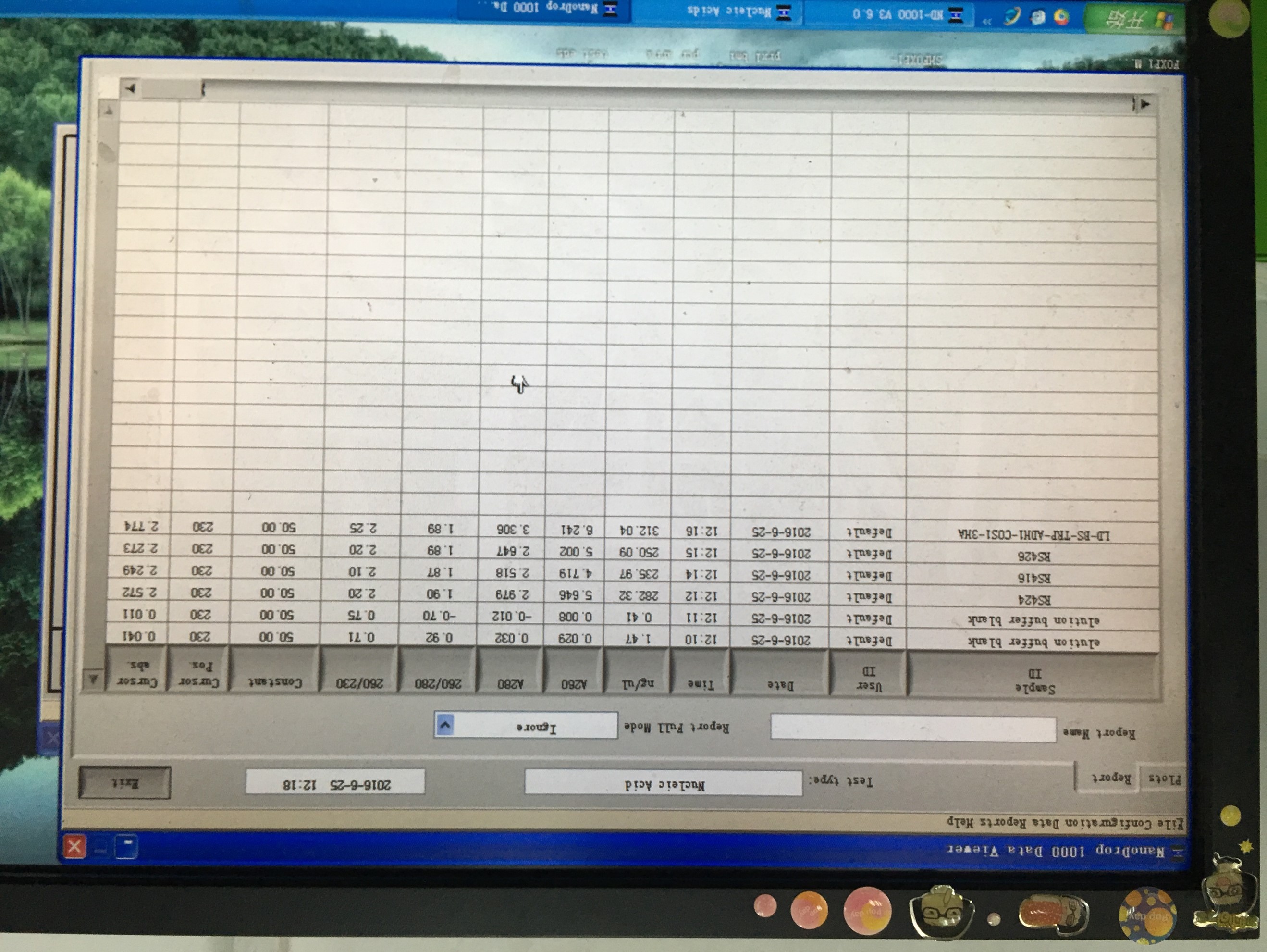
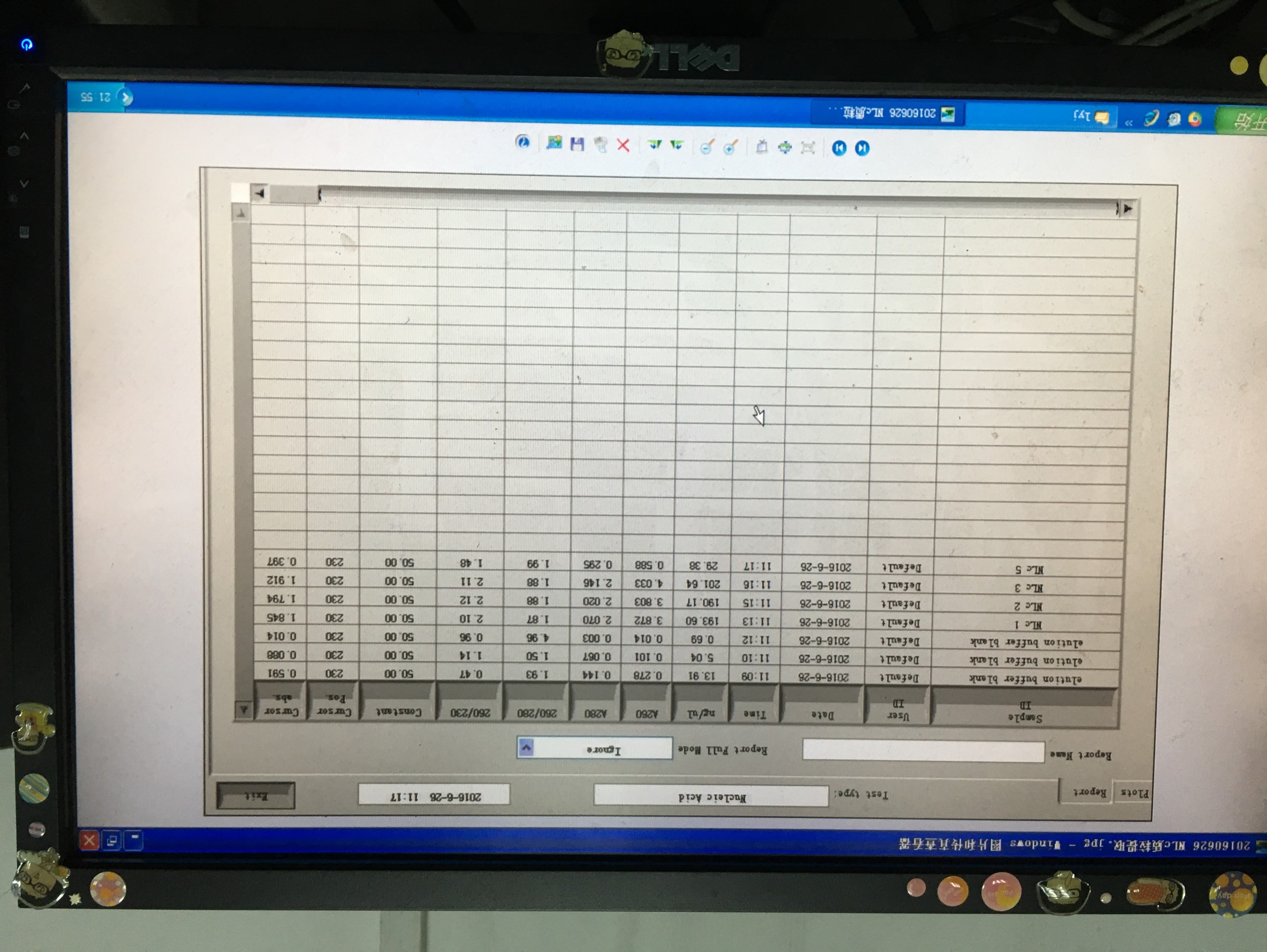
20160626
- Plasmid extraction of pcDNA3-Nanolantern
- ClaI-AvrII restriction digestion of LD-BS-ADH1-COS1
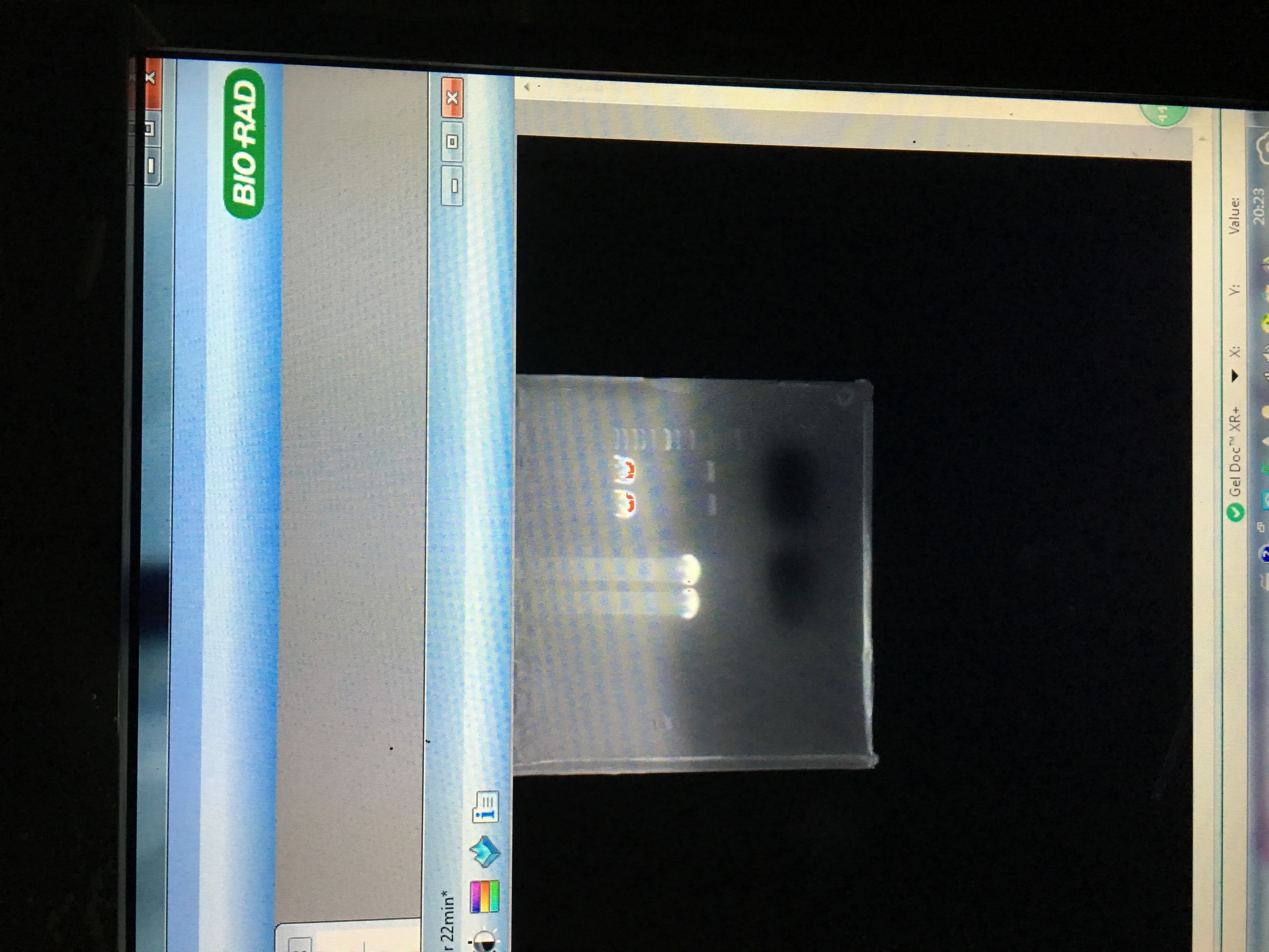
- PCR amplification of Nano-lantern cAMP 1.6 from pcDNA3-Nanolantern
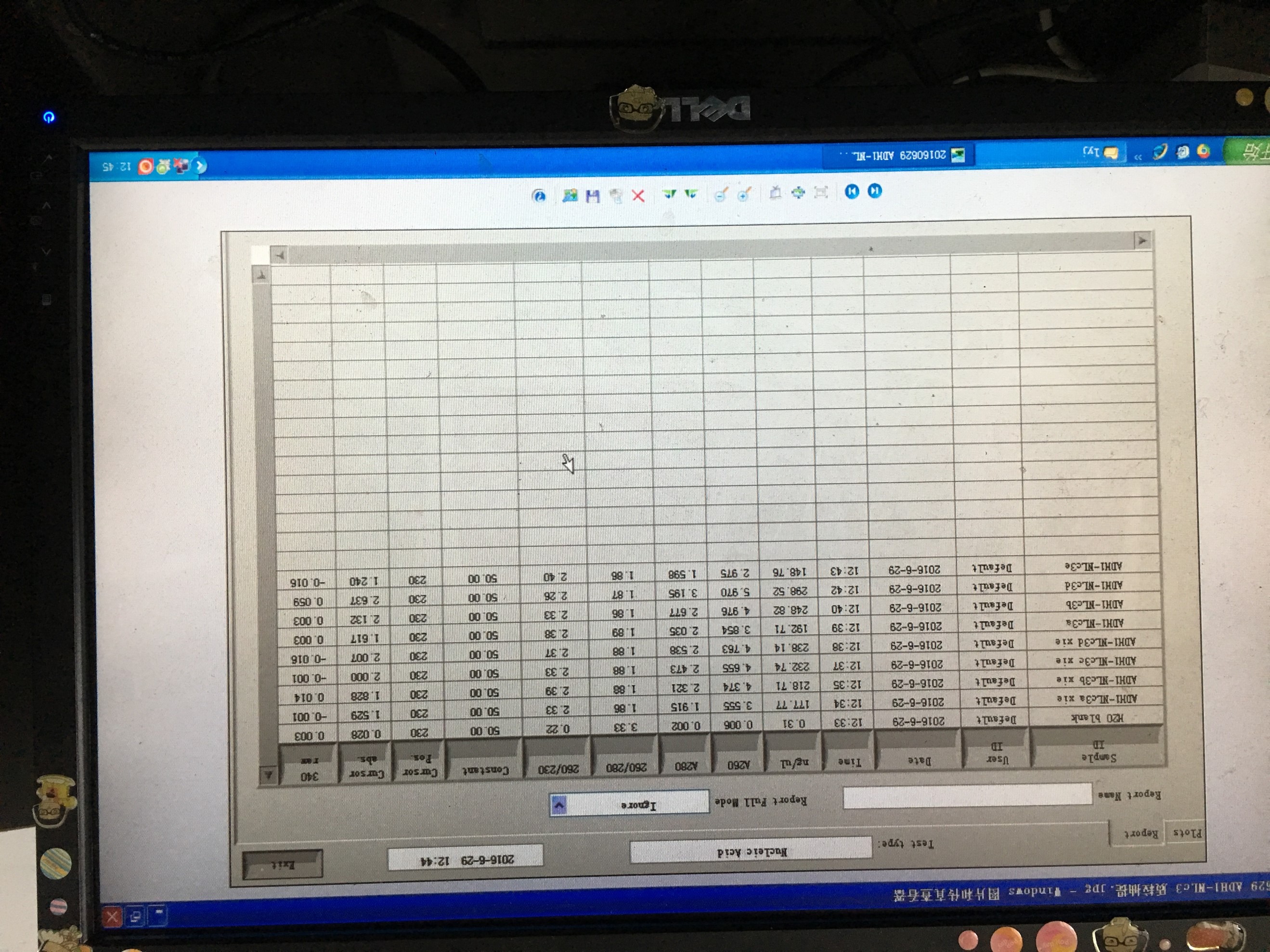
20160629
- Plasmid extraction of TRP-pADH1-NLc
- PCR amplify the GAL1 and CUP1 promoter from yeast genomic DNA and cloned them into plasmid backbones.
20160630
- Pick up colonies of GAL1 and CUP1 promoter with ADH1 terminator (TRP-CU/TRP-GAL).
20160701
- Plasmid extraction of TRP-CU/TRP-GAL.-
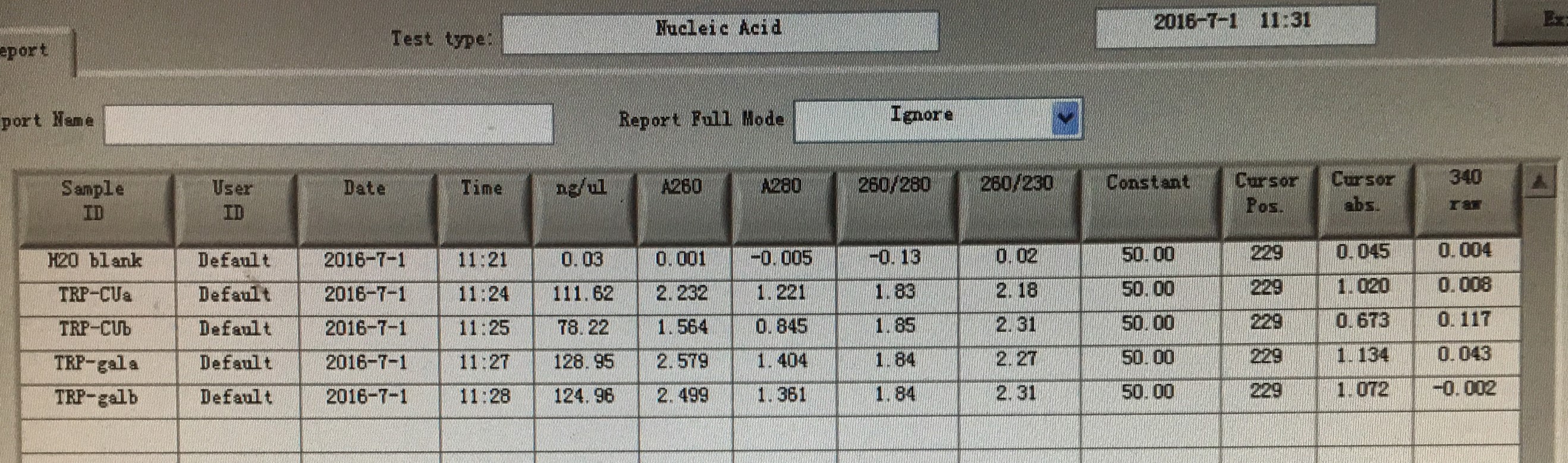
20160706
- Got strain DJ03 and 9060 from professor Zhiping Xie’s lab.
- Ordered coelenterazine-h from YEASEN BIOTECHNOLOGY.
20160707
- Transformation of DJ03 and 9060 with construct TRP-ADH-Nlc.
| Volume | OD600 | Control/Experimetnal | Digested DNA |
| DJ03 | 3ml | 0.9 | 1ml/2ml | 0μl/10μl |
| 9060 | 3ml | 0.8 | 1ml/2ml | 0μl/10μl |
20160709
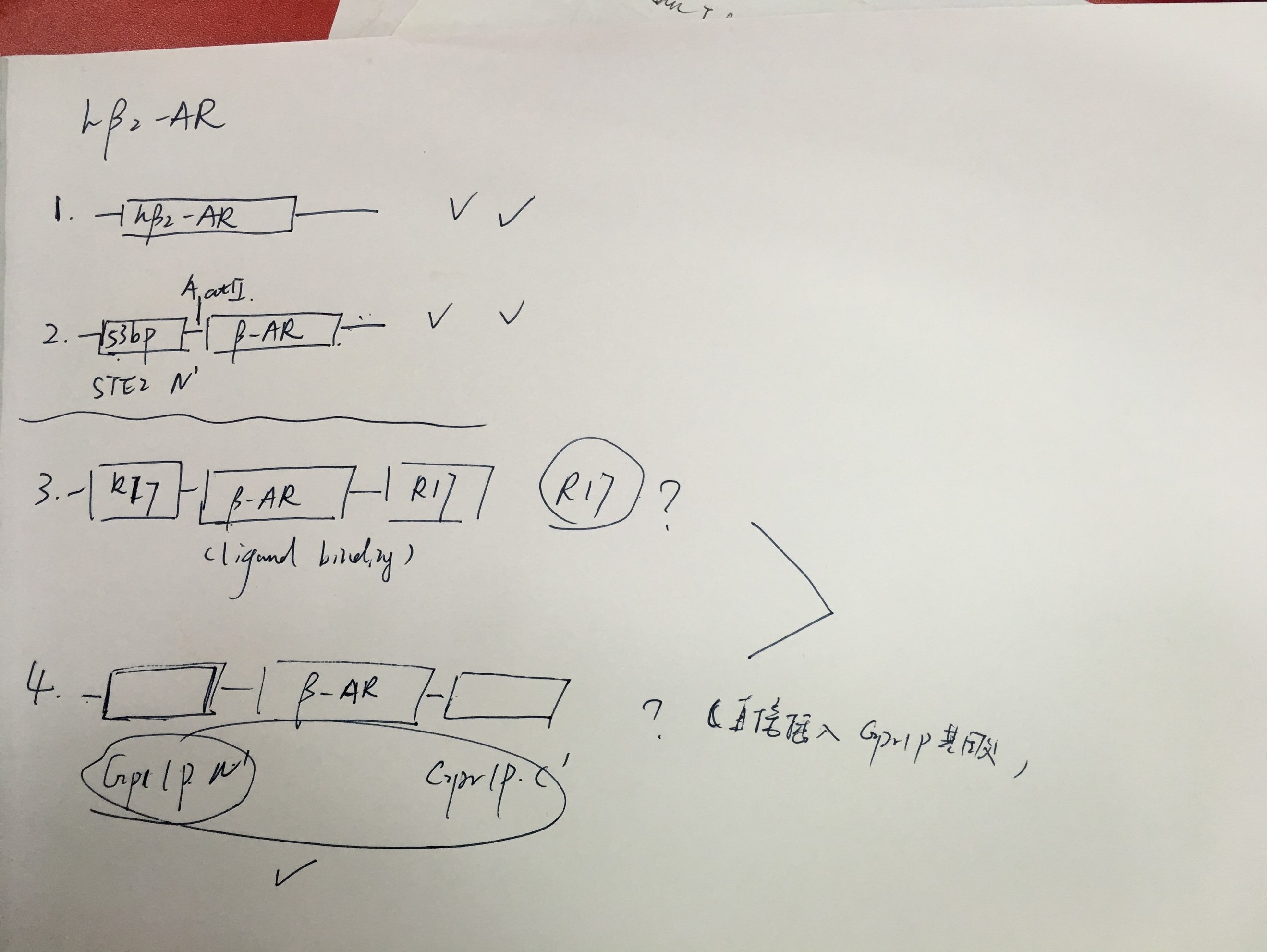
- Seeking for the cds for human adrenergic receptor beta 2 (ADRB2)
20160710
- Making modification plans for heterologous expression of ADRB2 in yeast.
20160711~20160713
- Finishing the construct TRP-CU-NLc and TRP-CU-NLc
20160713
- Fluorescent microscopy of TRP-ADH1-NLc to detect the YFP signal
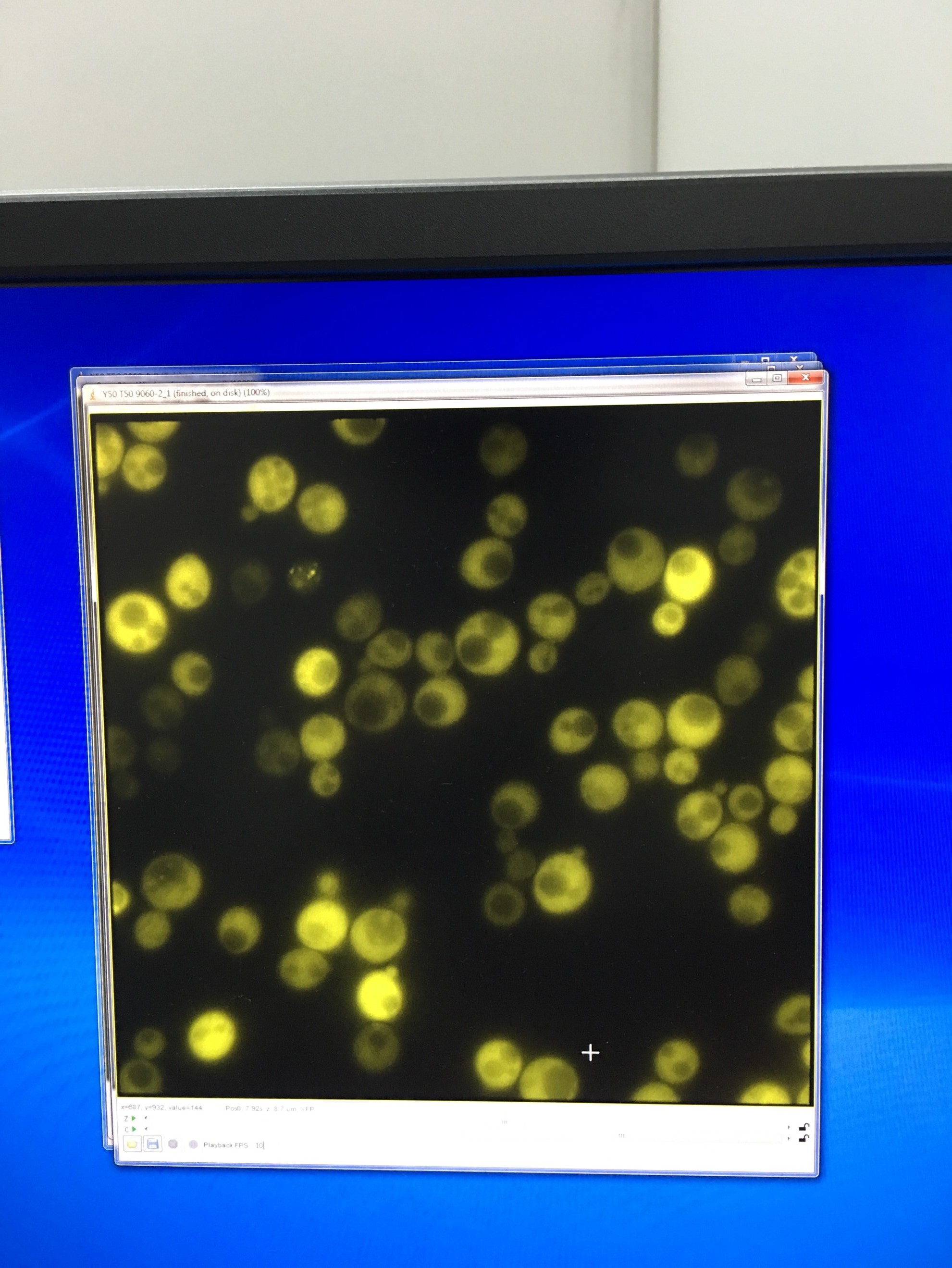
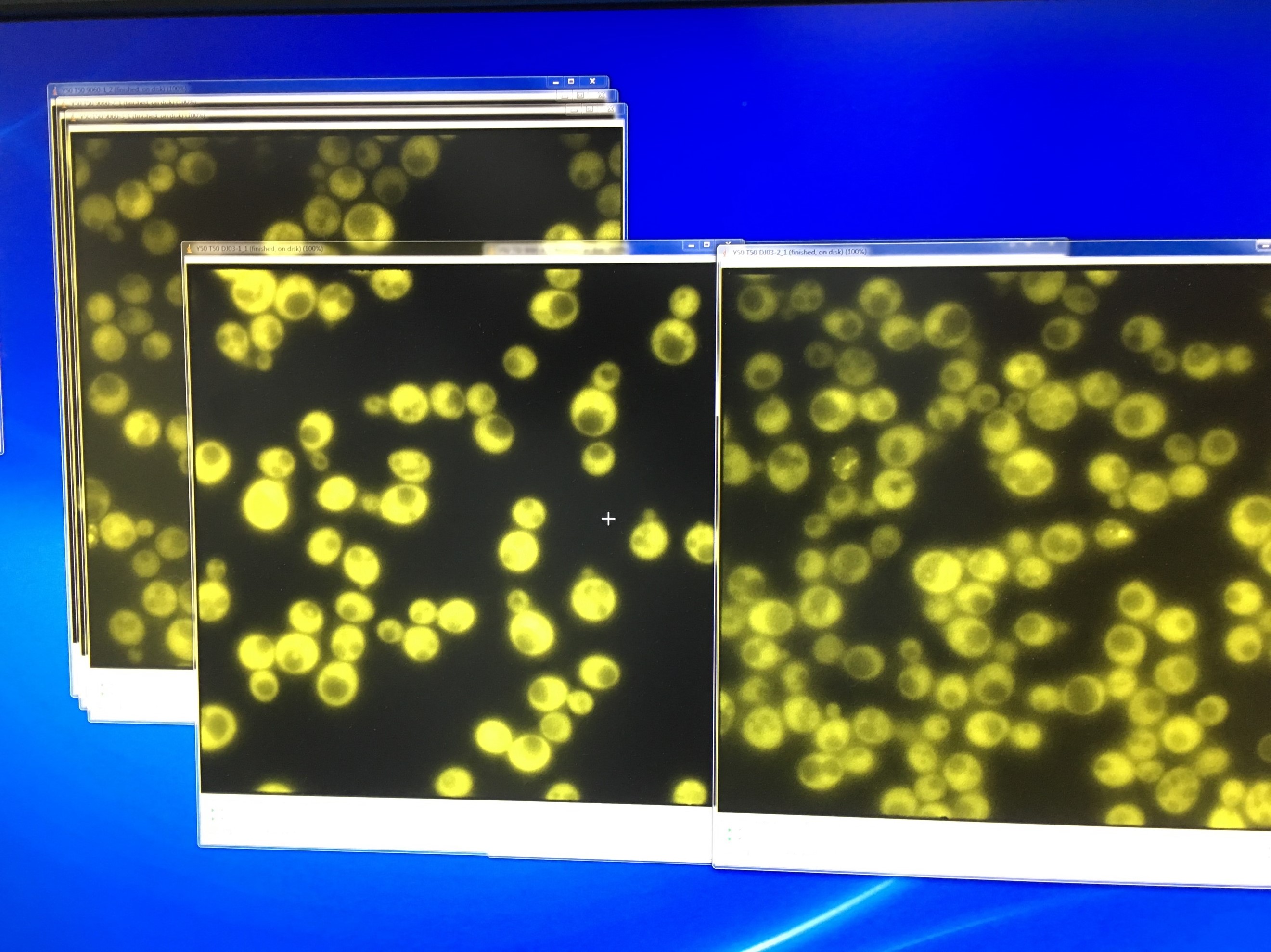
20160714
- Colony PCR of TRP-GAL-NLc3 abc/TRP-CU-NLc3 abc
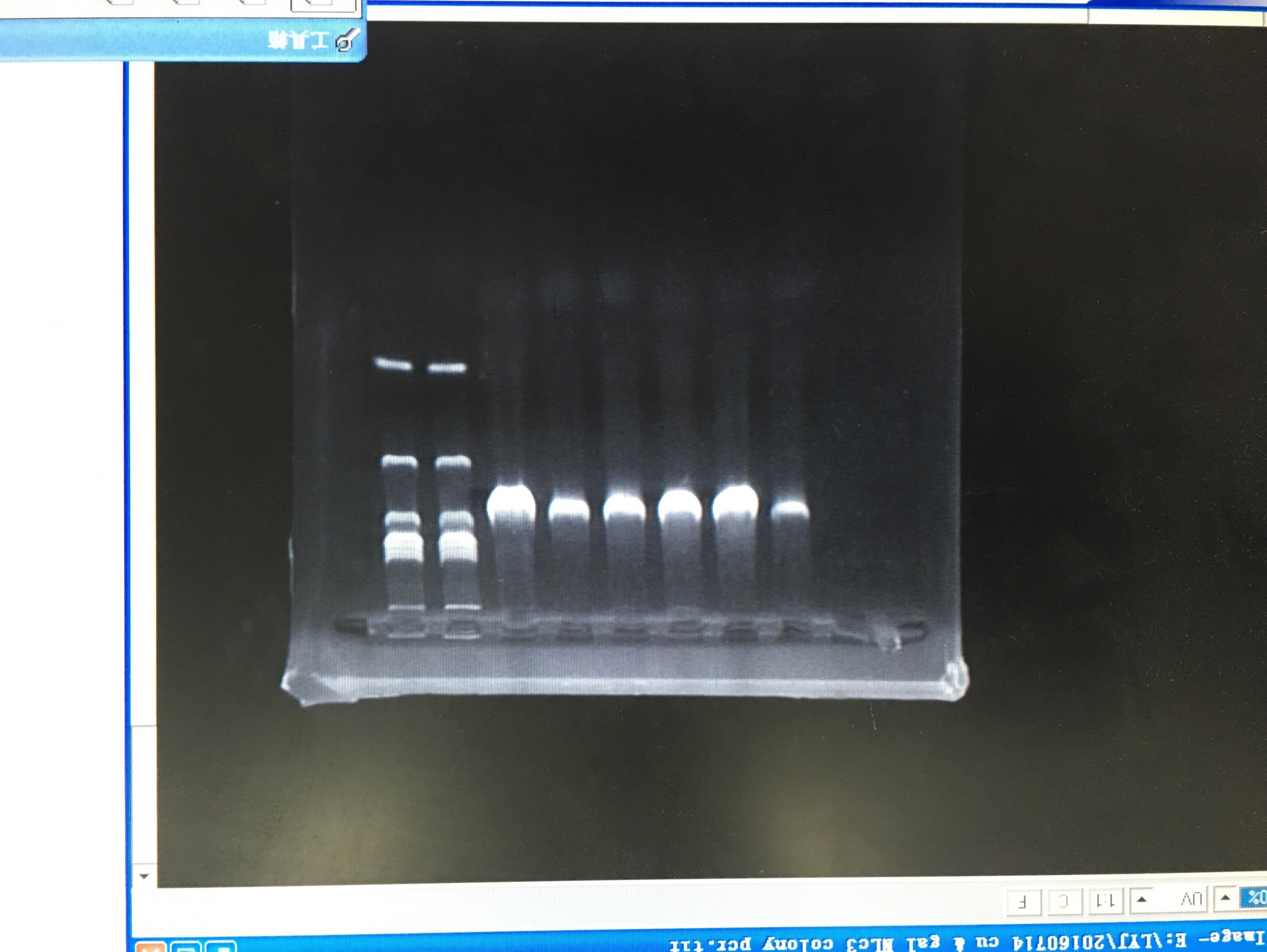
- Plasmid extraction of TRP-GAL-NLc3 abc/TRP-CU-NLc3 abc
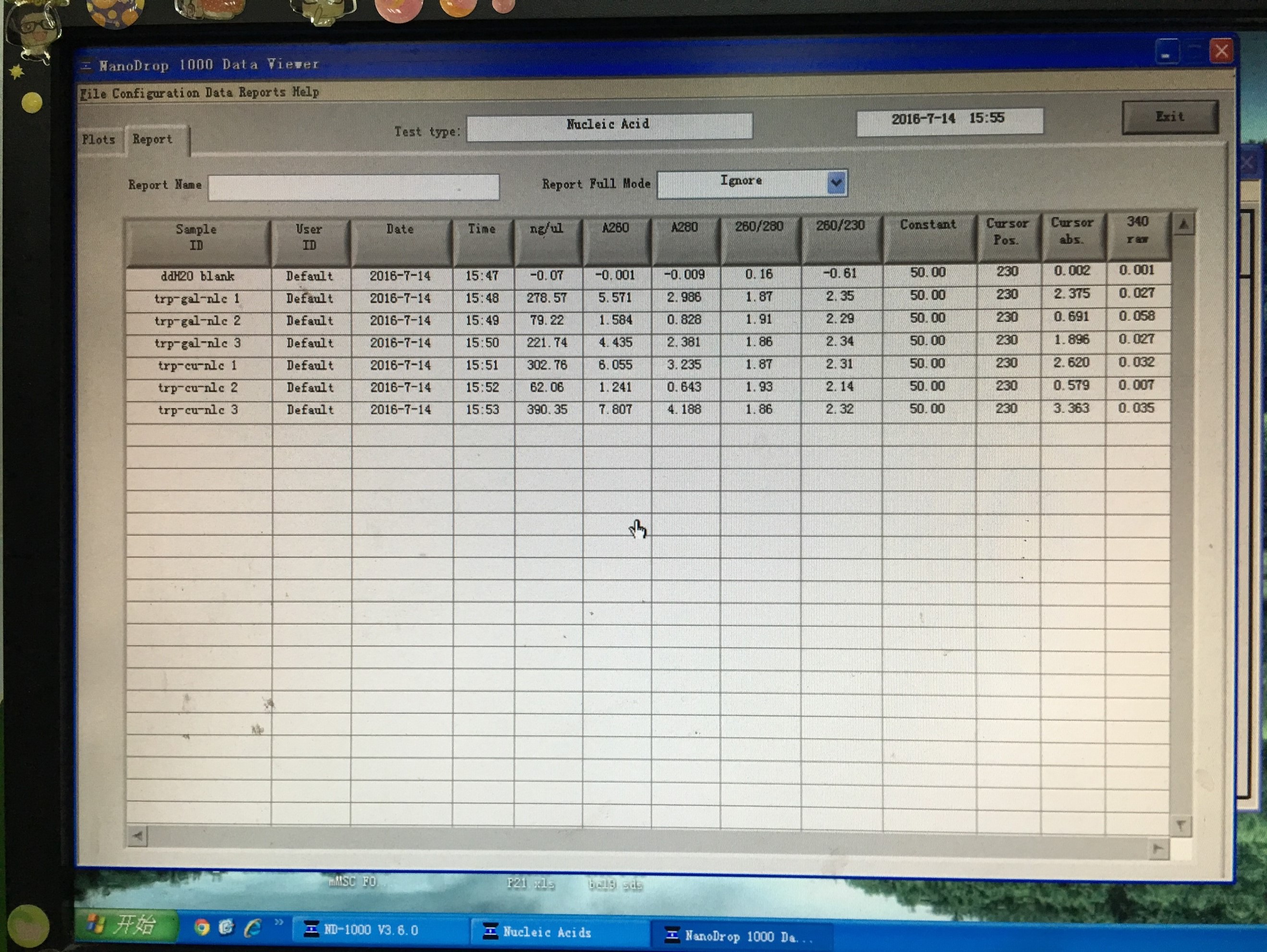
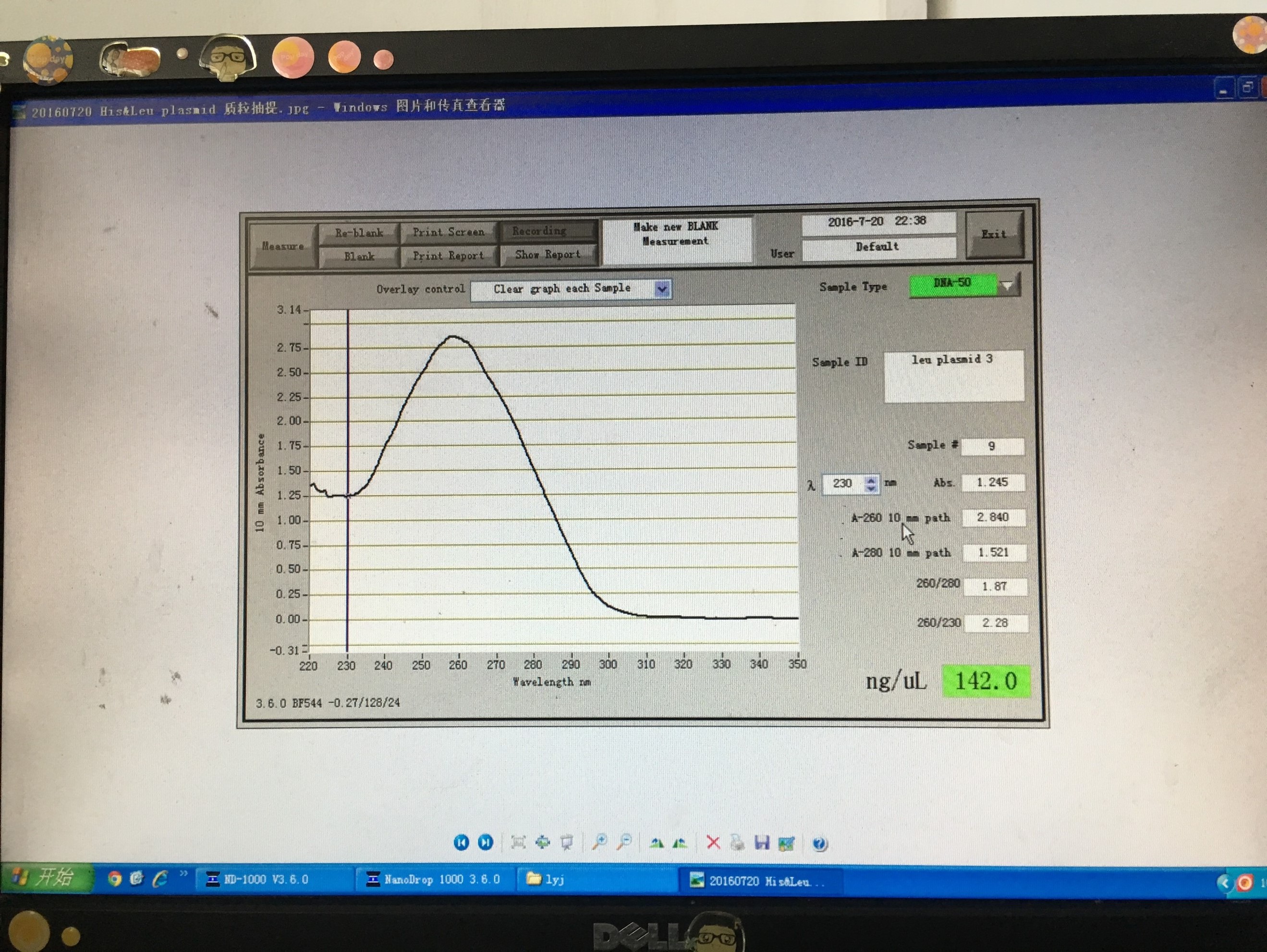
20160719~20160720
- Obtained plasmid backbone LD-BS-HIS3 and LD-BS-LEU2 from Zhiping Xie’s lab and amplified them to make larger amount.
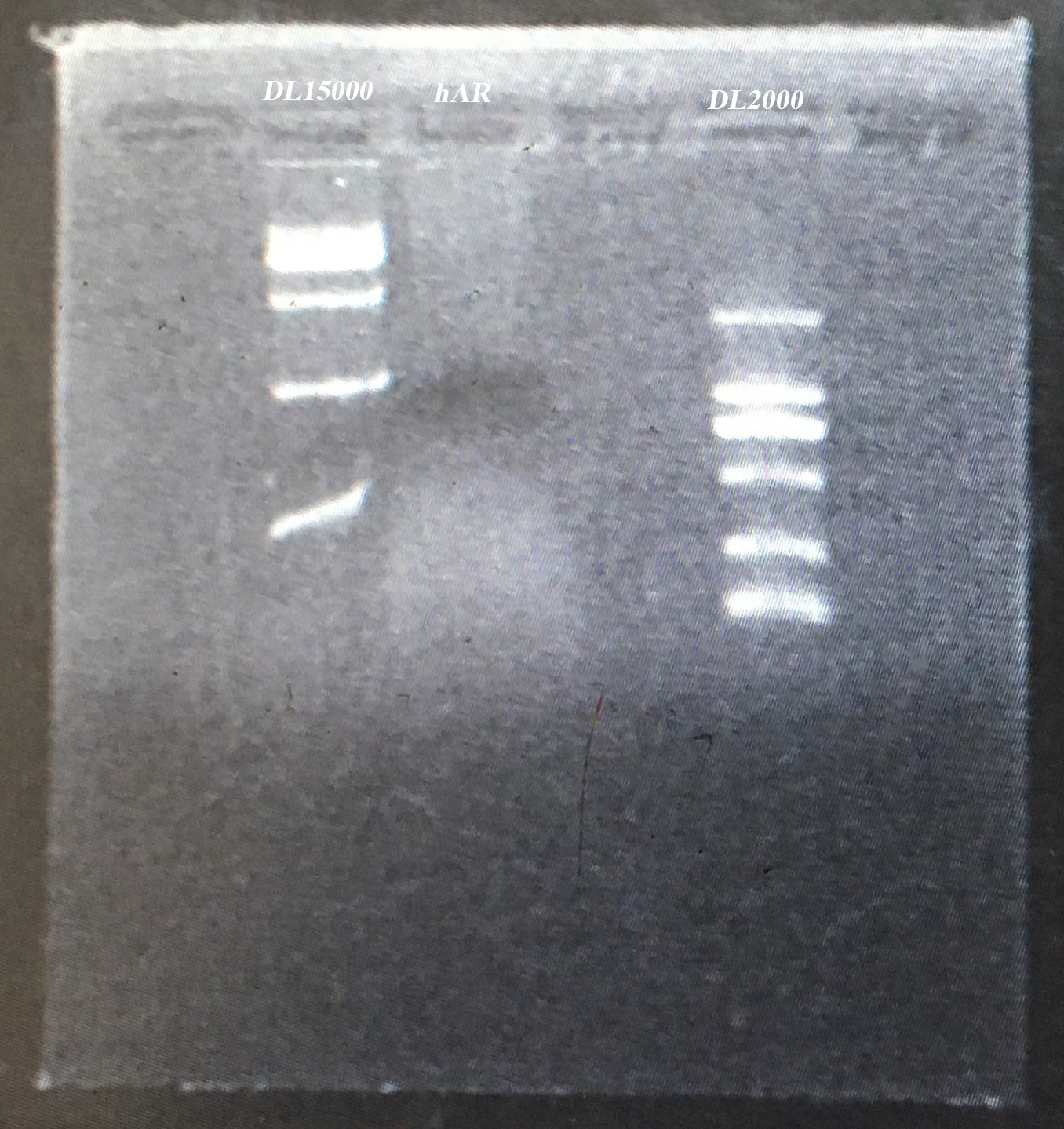
20160723
- Tried to amplify the ADBR2 cds from genomic DNA of A549 cells
20160726
- Amplify the ADRB2 gene from genomic DNA of A549 cells using different melting temperature and different polymerase
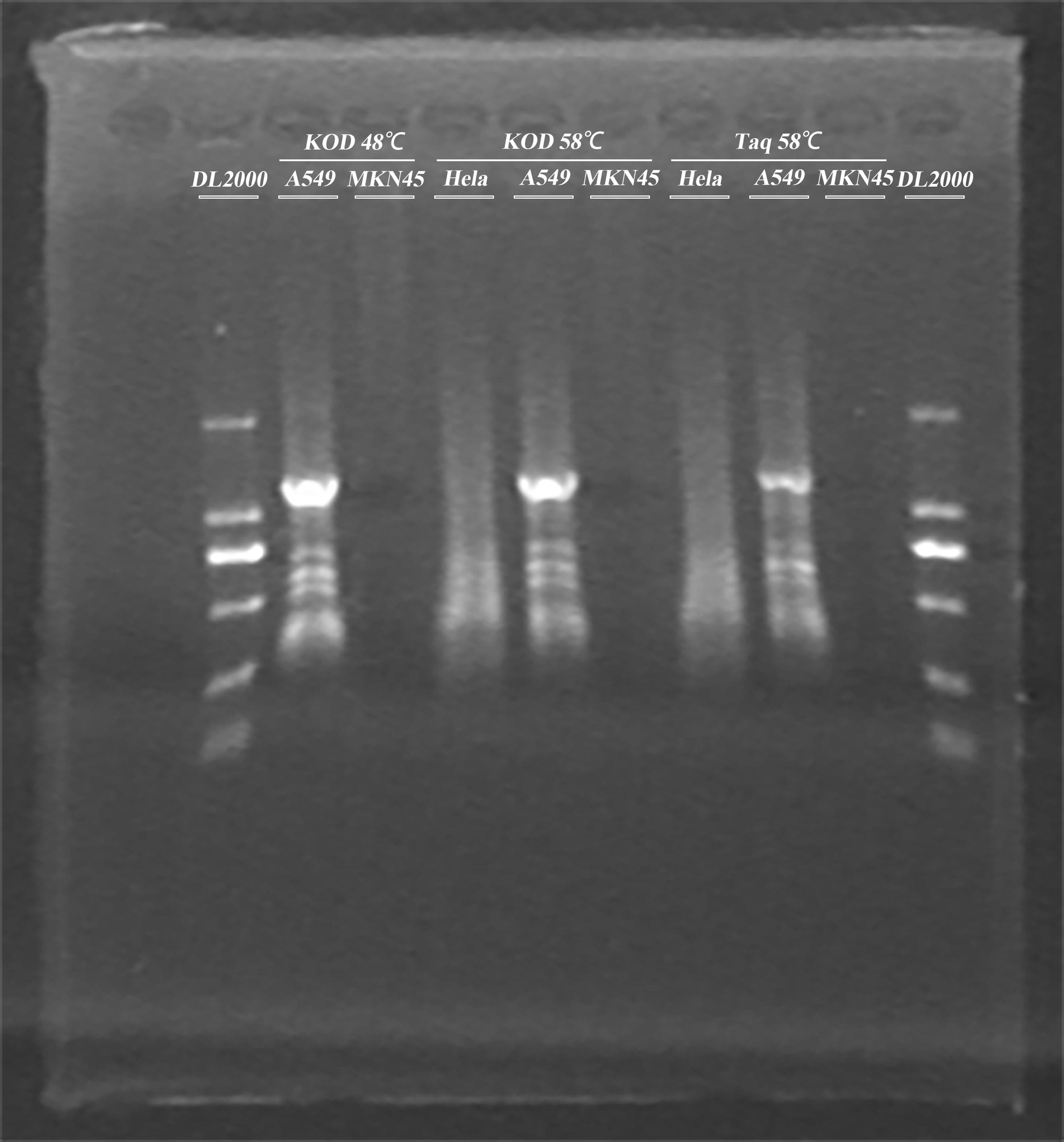
- Gel extortion of ADRB2 gene
- Enzyme digestion of LD-BS-TRP-pADH1-CDS1-3HA and LD-BS-TRP-GAL-3HA-DGA1-aid-6HA
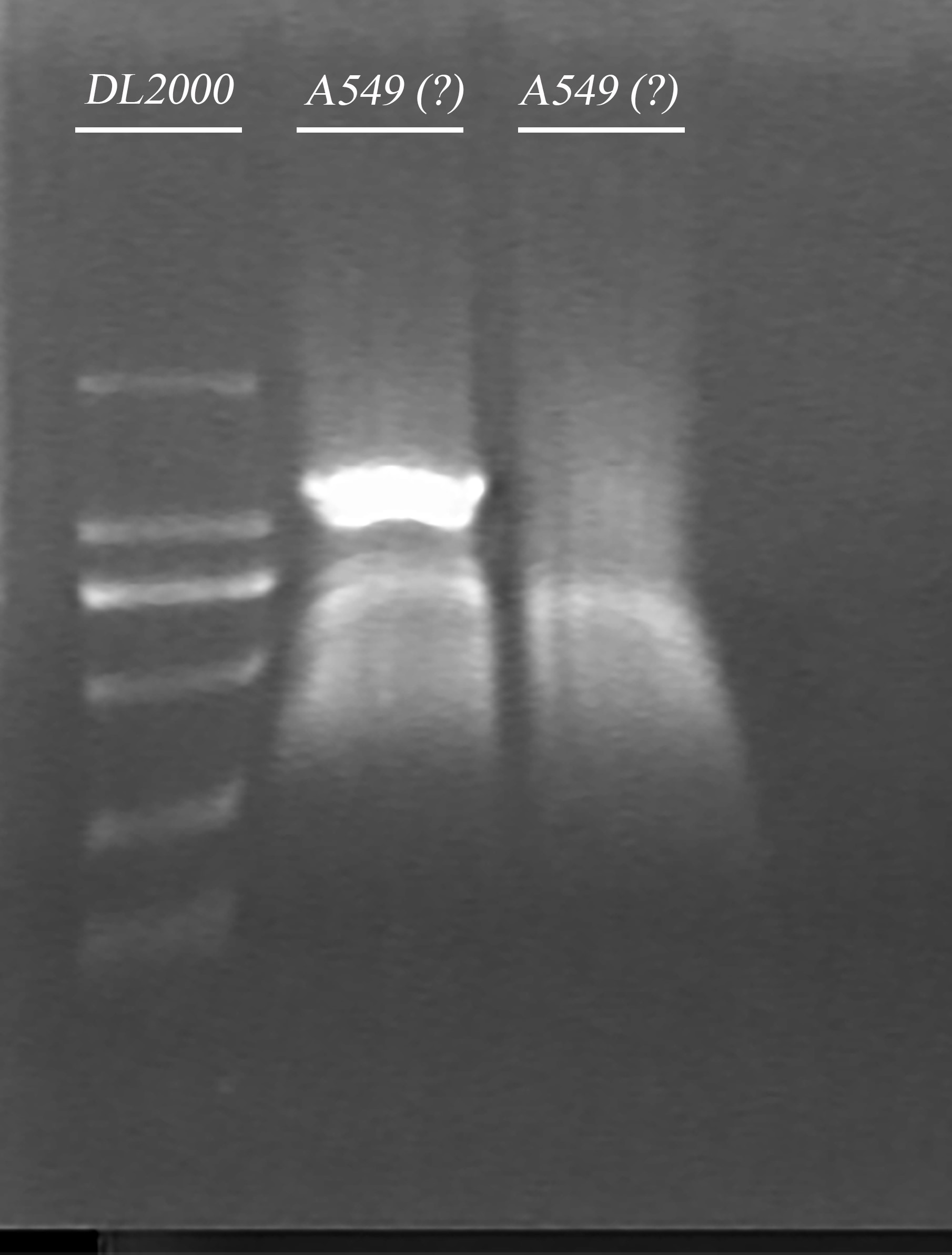
- Amplify the ADRB2 gene from the fragment from gel extraction product
| Reagents | Volume |
| KOD plus neo | 1 μl |
| 10x Buffer for KOD -Plus- Neo | 5 μl |
| 2mM dNTPs | 5 μl |
| 25mM MgSO4 | 3 μl |
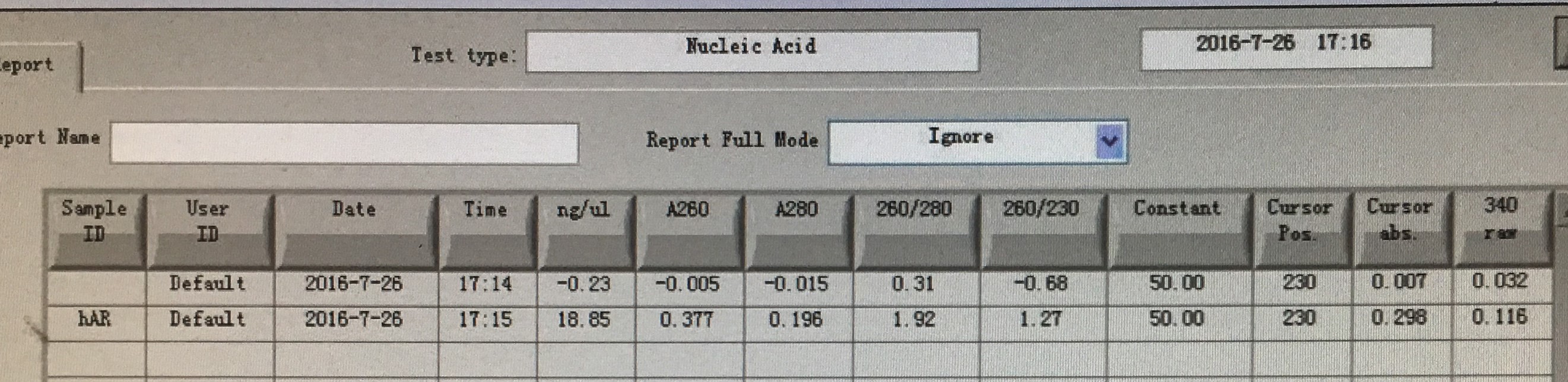
| hAR | 10 μl |
| 10 pmol/µl Primer | 1x2 μl |
| ddH2O | 24 μl |
Primer:
hAR : hAR-R/hAR-F
TRP-ADH-hAR : TRP-ADH-hAR-R/ TRP-ADH-hAR-F
TRP-GAL-hAR : TRP-GAL-hAR-R/ TRP-GAL-hAR-F
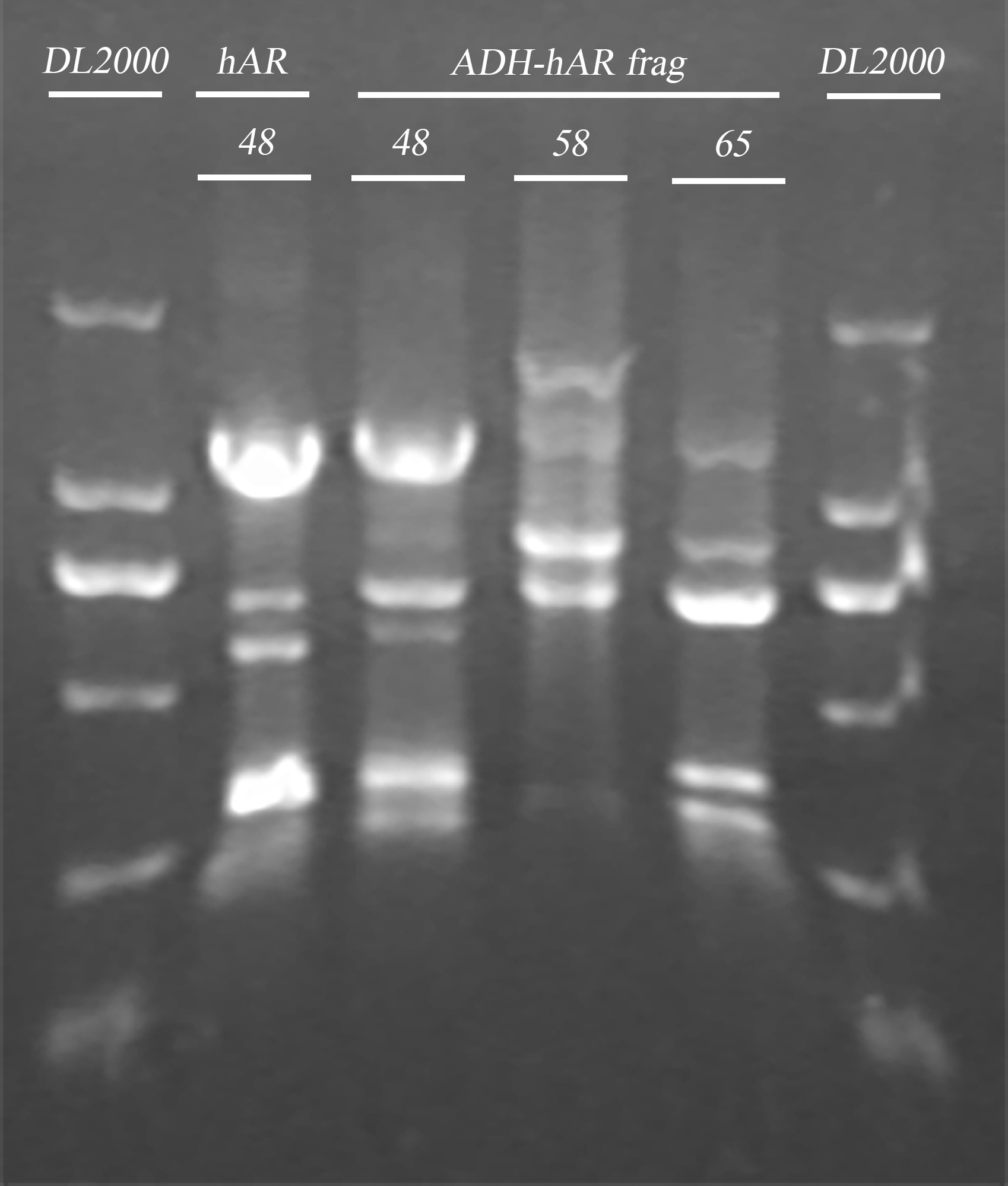
- Gel electrophoresis of the PCR product in 5 and endonuclease digested product in 4
- Gel extraction of endonuclease digested product
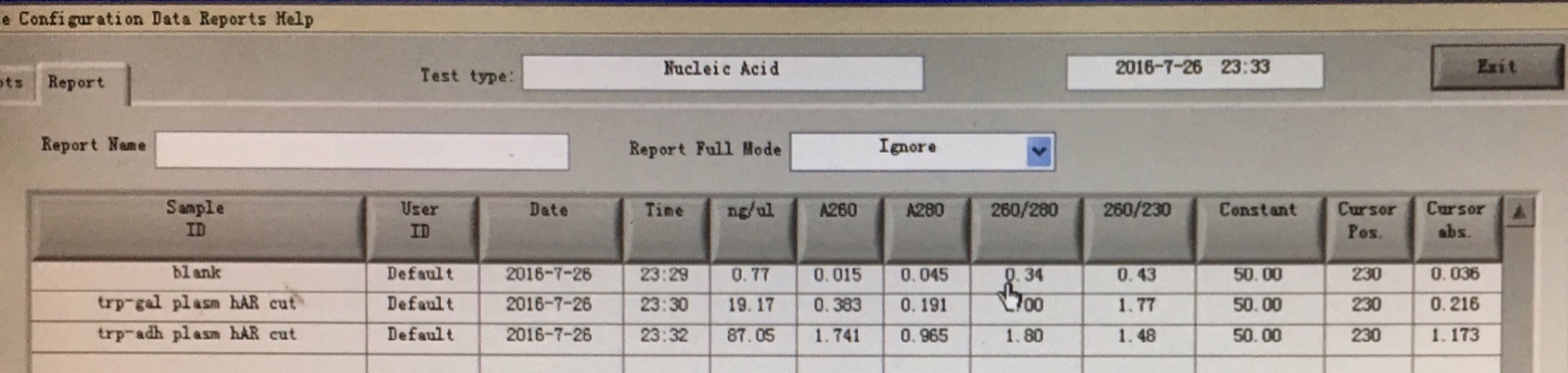
20160727
- PCR amplification of ADRB2 gene.
450px
- PCR cleaning
- Gel electrophoresis of product in 3.
- Gel extraction
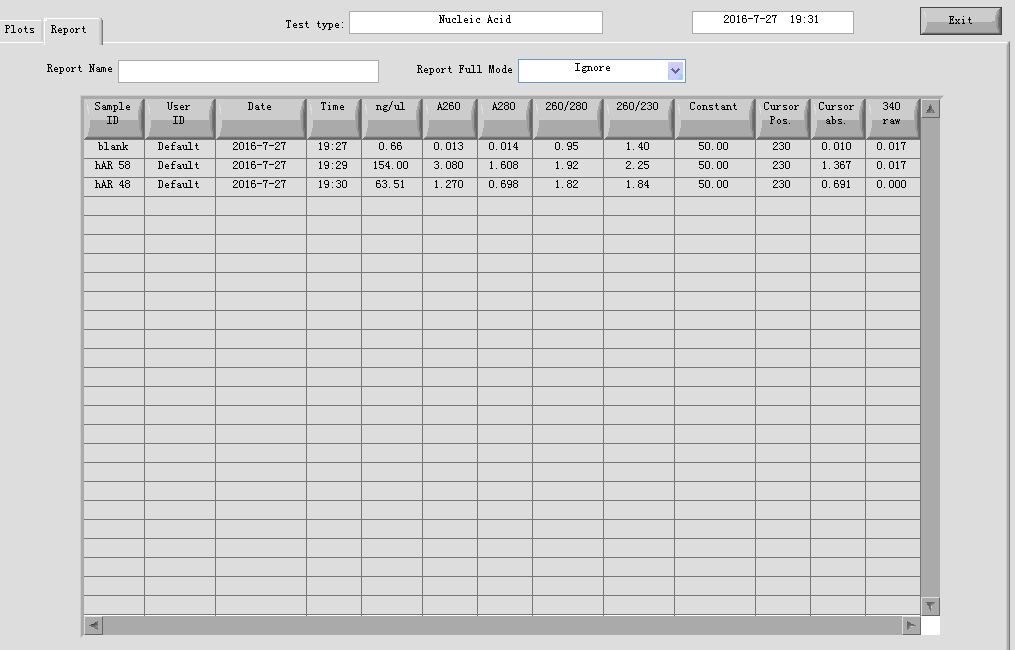
20160728
- ADRB2 gene fragment PCR
| Name | Primer | Tm | Template(μl) | ddH2O (μl) | Total(μl) |
| hAR | hAR-R/hAR-F | 48℃ | (48℃) 2.4 | 11.2 | 20 |
| ADH-hAR | TRP-ADH-hAR-R/ TRP-ADH-hAR-F | 48℃ | (48℃) 6 | 28 | 50 |
| ADH-hAR | | 58℃ | (58℃) 3 | 31 | 50 |
| ADH-hAR | | 65℃ | (48℃) 6 | 28 | 50 |
Cycle:
| Name | Temperature | Duration |
| Pre-denaturation | 95℃ | 3 min |
| Denaturation | 95℃ | 40s | 39x |
| Anealing | 48/58/65℃ | 30s | |
| Extension | 68℃ | 1min 30s | |
| Final extension | 72℃ | 10min |
|
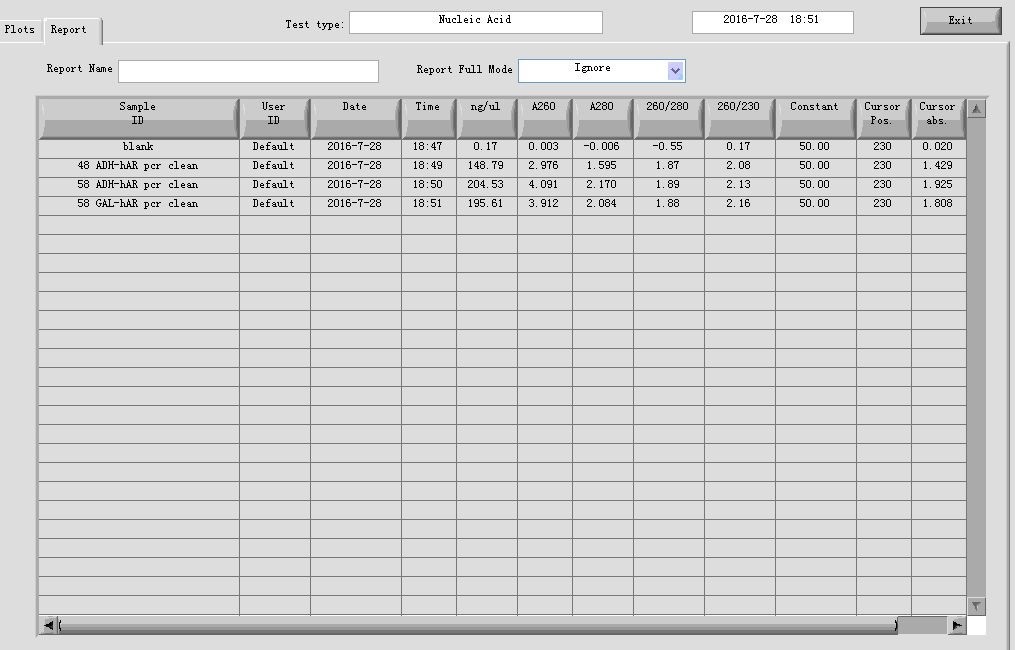
- PCR cleaning
Mixed the parallel groups in step 1, then added equal volume of Buffer CP to them, and follow the standardized protocol provided by the manufacturer. The final elution volume is 30 μl.
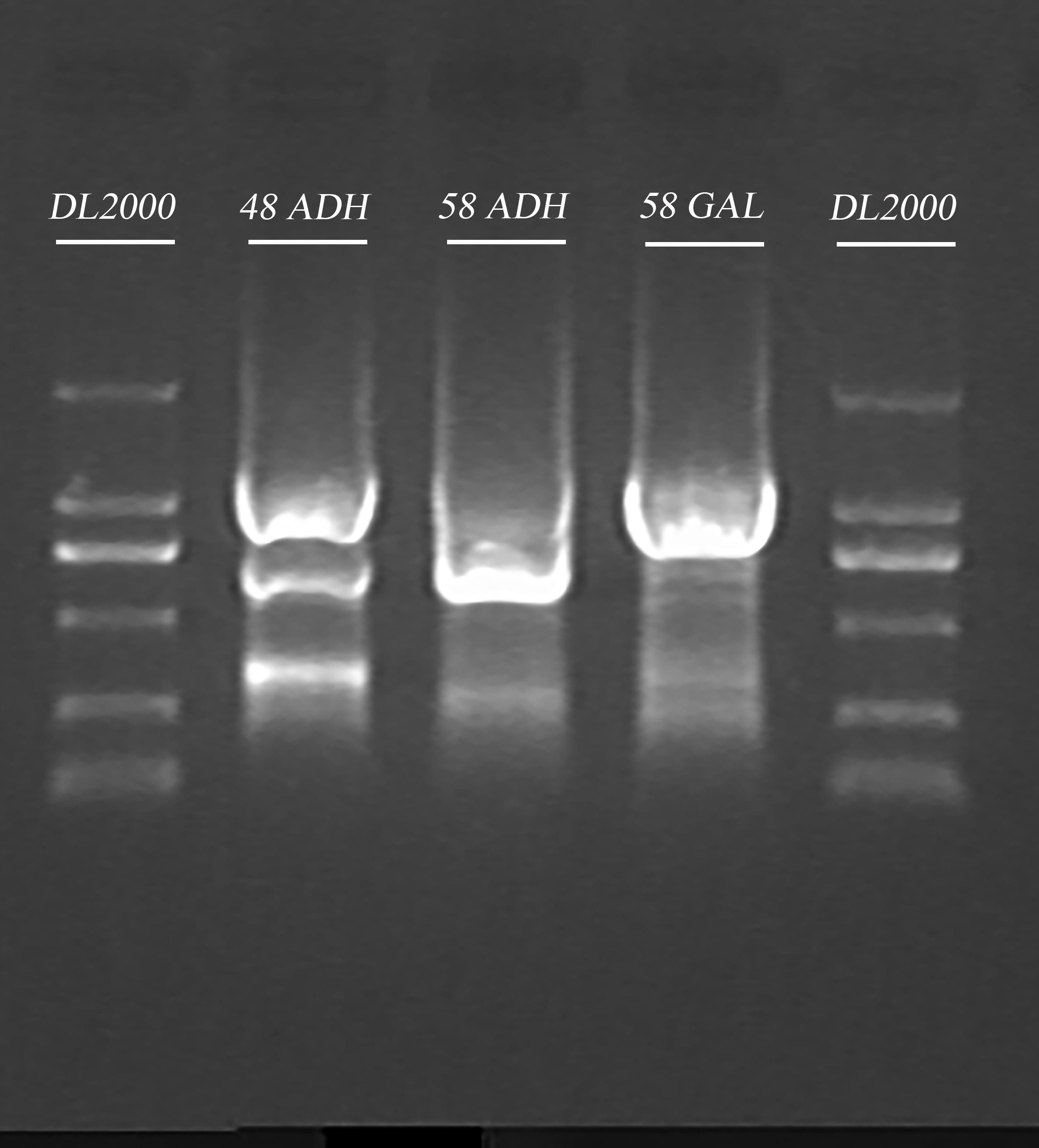
- Gel recovery
Ran the samples obtained in Step.3 and recovered the gel. The elution volume is 30 μl.
| Gel | DNA sample volume (after PCR cleaning) | 6 x Loading buffer | DL2000 |
| 1% agarose | 30 μl | 6 μl | 10 μl |
|
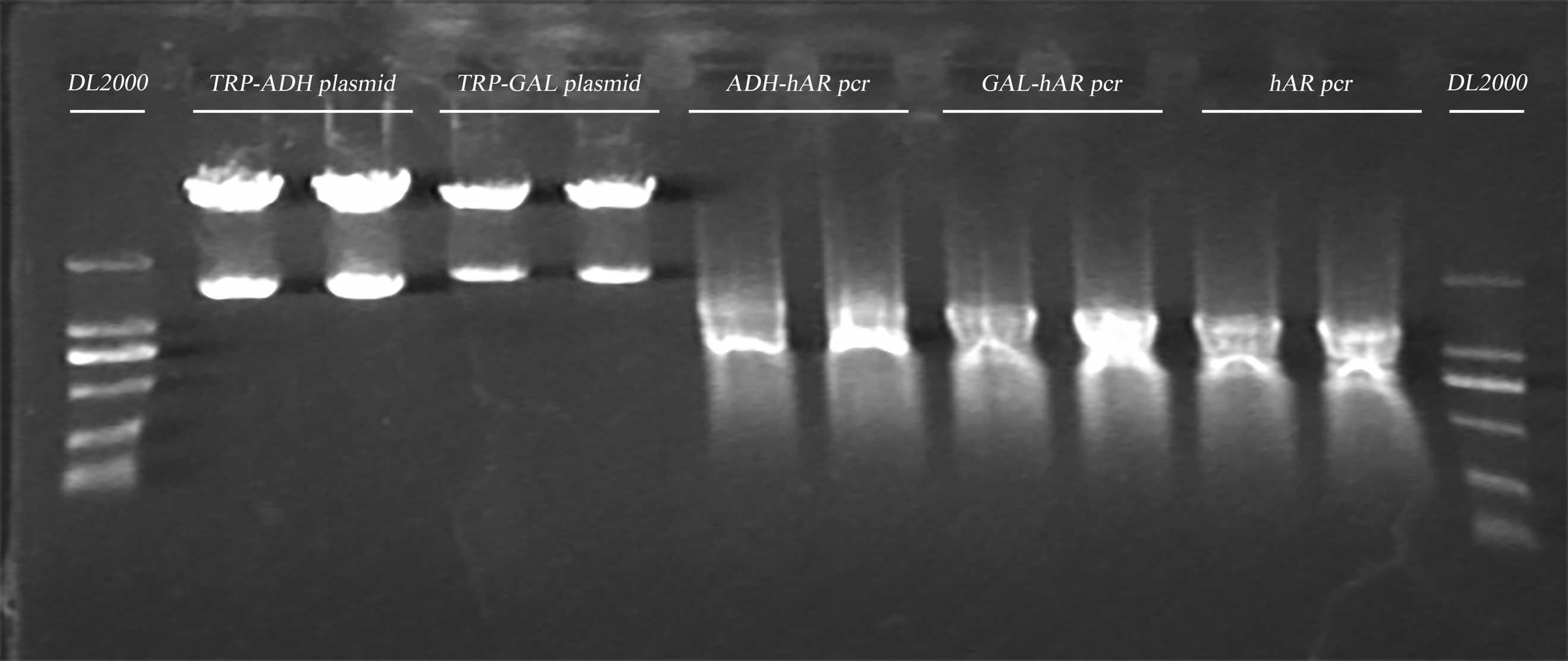
- Insert the fragment into the plasmid TRP-ADH/TRP-GAL
The ligation was achieved using ClonExpress II One Step Cloning Kit following the manufacturer’s protocol.
- Transformation of DH5a competent cells
| DNA volume | Competent cells | LB media | Spread on plate |
| 10 μl | 100 μl | 600 μl | 350 μl |
|
20160729
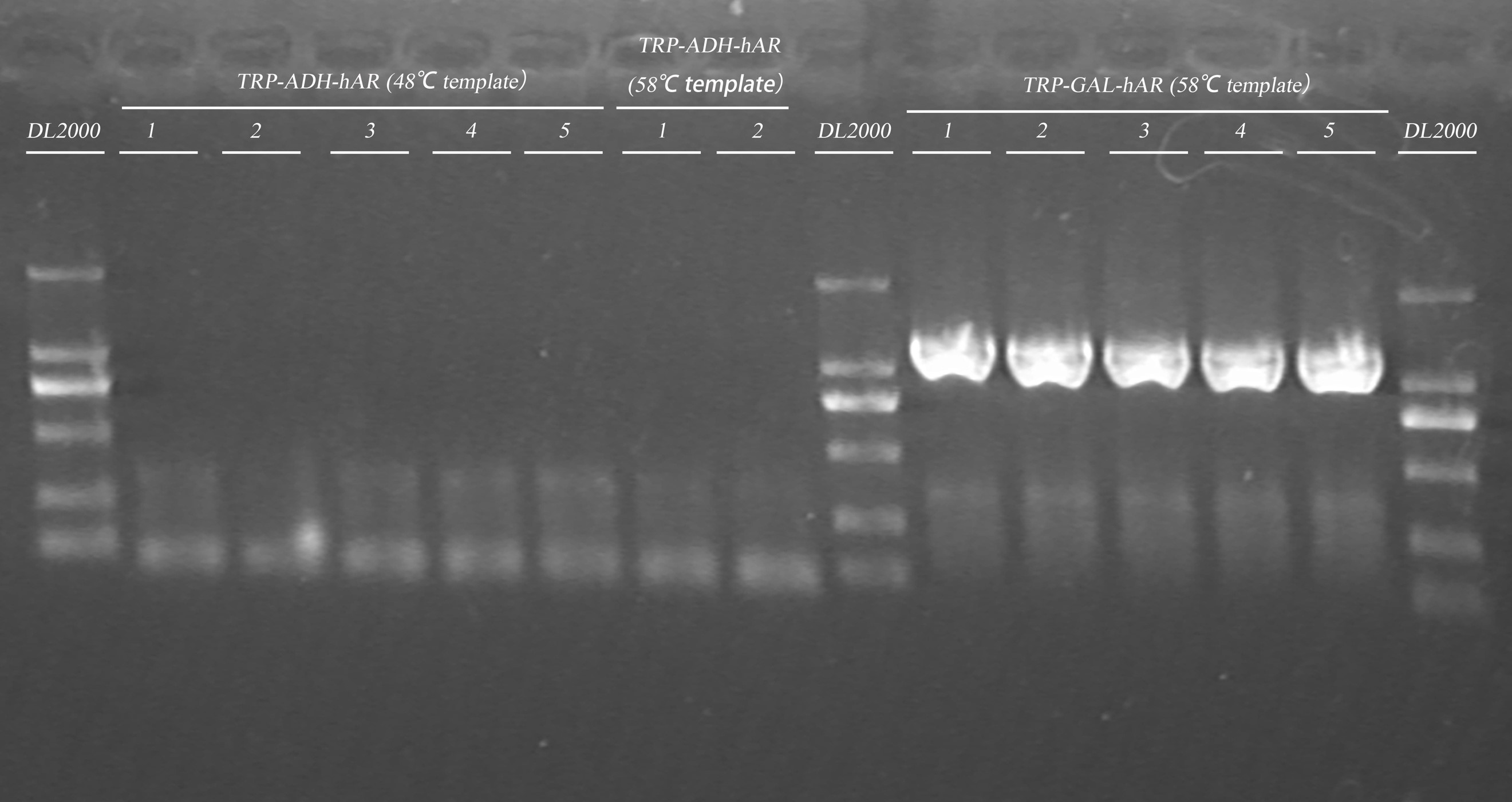
- Pick up positive colonies
| Strains | Positive colonies |
| TRP-ADH-hAR (48) | 5/5 |
| TRP-ADH-hAR (58) | 2/2 |
| TRP-GAL-hAR (58) | 5/N |
20160730
- Colony PCR
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Culture | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:
hAR : hAR-R/hAR-F
TRP-ADH-hAR : TRP-ADH-hAR-R/ TRP-ADH-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 35x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
- Gel electrophoresis of PCR products in Step.1
| Gel | PCR product | 6 x Loading buffer | DNA ladder |
| 1% agarose | 20μl | 4μl | 5μl |
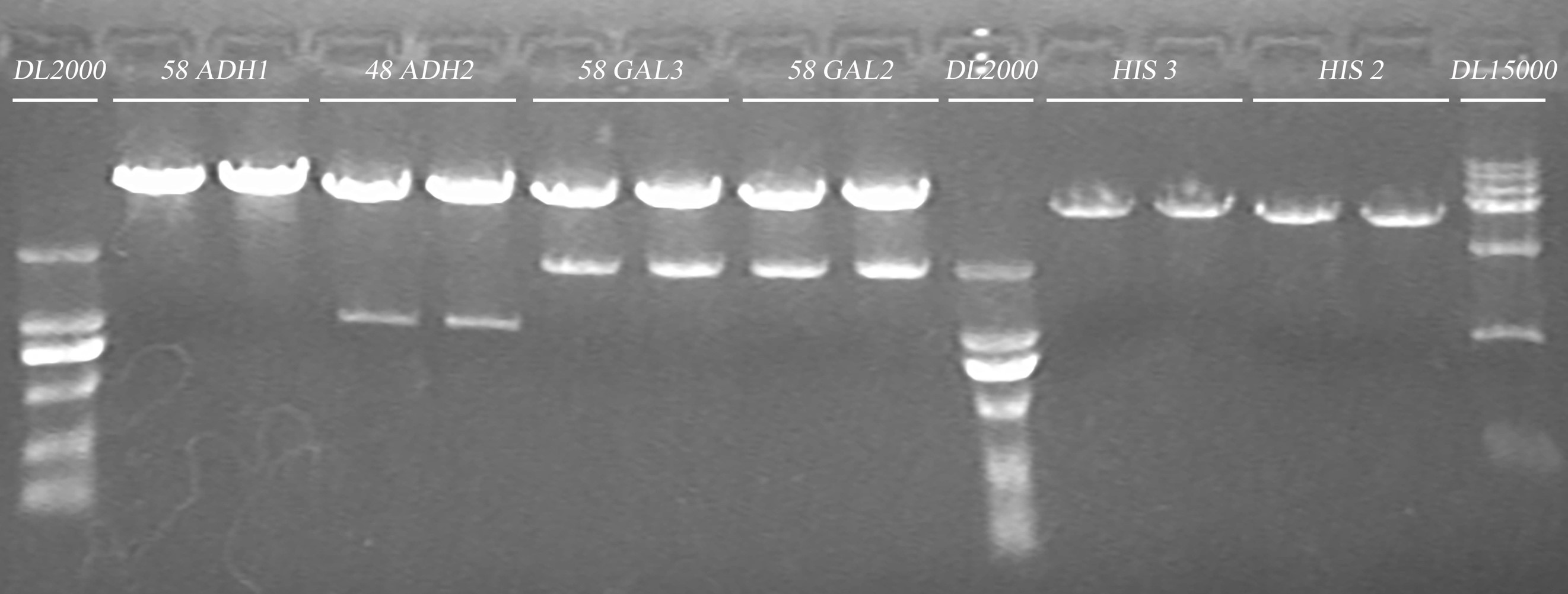
- Plasmid extraction
58GAL1,2,3,4,5/48ADH1,2/58ADH1
Elution: 30μl in ddH2O;
- Endonuclease digestion
| Plasmid | 12μl |
| Endonuclease | 1μl HindIII (HF) |
| 1μl ApaI |
| Buffer | 5μl Cutsmart |
| ddH2O | 31μl |
Gel electrophoresis
| Gel | Digestion product | 6 x Loading buffer | DL2000/15000 |
| 1% agarose | 50μl | 10μl | 10μl |
- Gel recovery
Elution: 15μl in ddH2O
20160731
- Ligation
Ligation process were done followed the standardized protocol from the manufacturer, the plasmids and DNA volume were set as follows:
| Construct | Plasmid | DNA | Total volume |
| HIS2-GAL2 | 6.7 μl HIS2 | 9 μl GAL2 | 20 μl |
| HIS3-GAL3 | 7.4 μl HIS3 | 9.6 μl GAL3 | 20 μl |
22℃ 1h
- Transformation of DH5a competent cells
| DNA volume | Competent cells | LB media | Spread on plate |
| 10 μl | 50 μl | 300 μl | 360 μl |
20160801
- colony pcr of HIS3-GAL3-hAR a/b/c +HIS2-GAL2-hAR a/b/c transformed cells
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Culture | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:TRP-GAL-hAR-R/ TRP-GAL-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 30x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
- Gel electrophoresis of PCR products in Step.1
| Gel | PCR product | 6 x Loading buffer | DNA ladder |
| 1% agarose | 20μl | 4μl | 5μl |
10μl/well
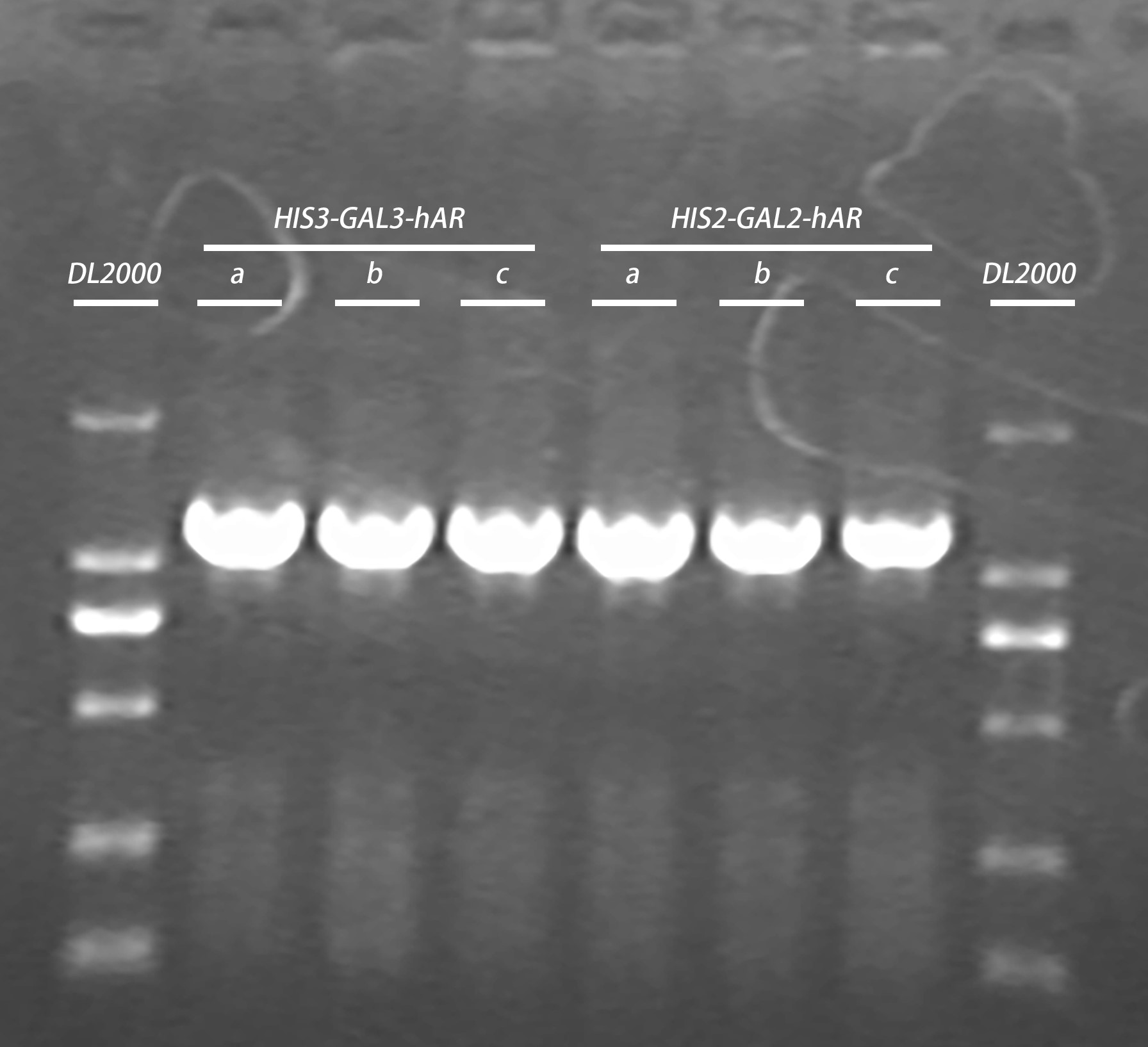
- HIS3-GAL3-hAR a/b/c +HIS2-GAL2-hAR a/b/c +TRP-ADH a/b plasmid extraction
20160804
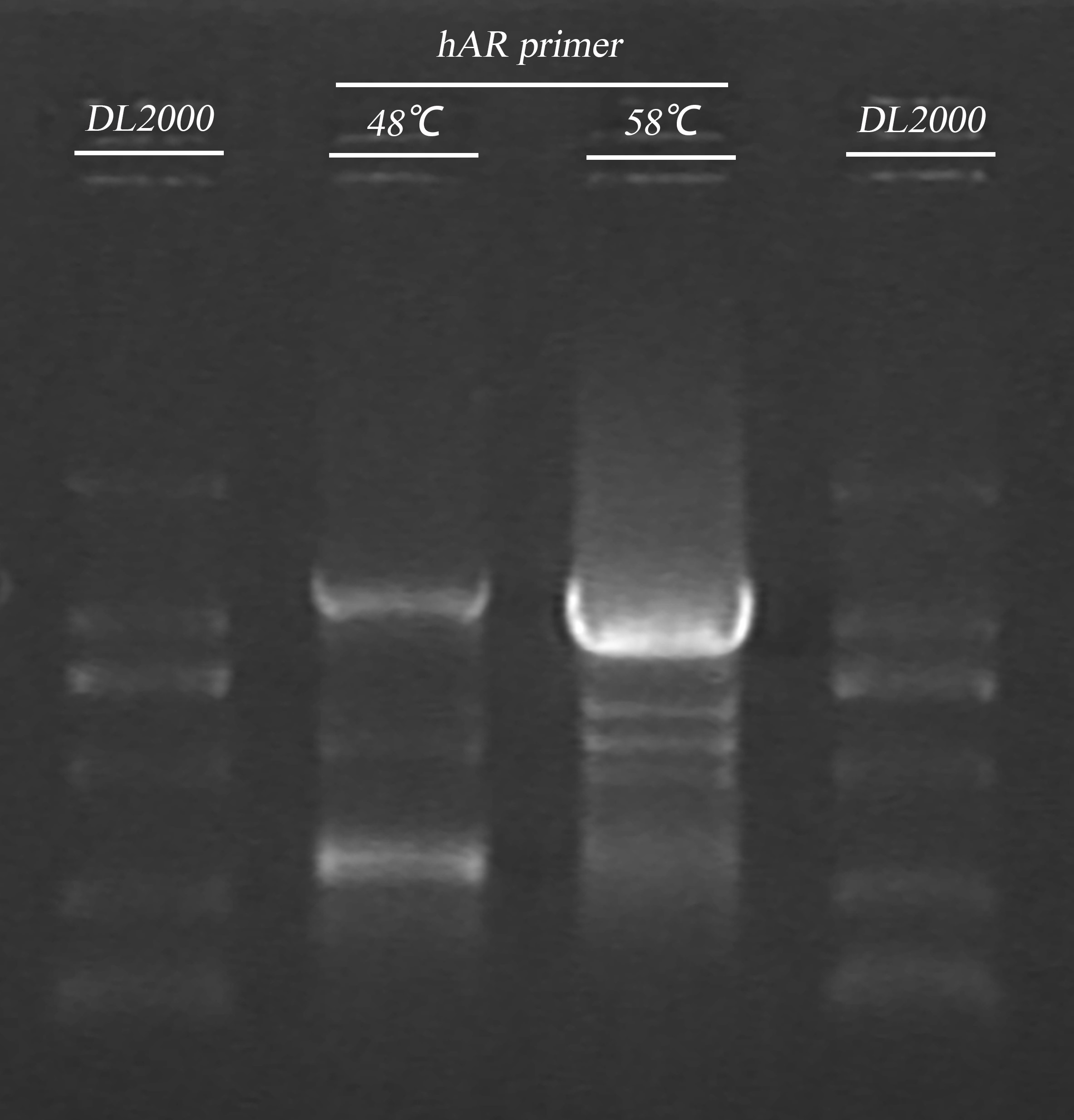
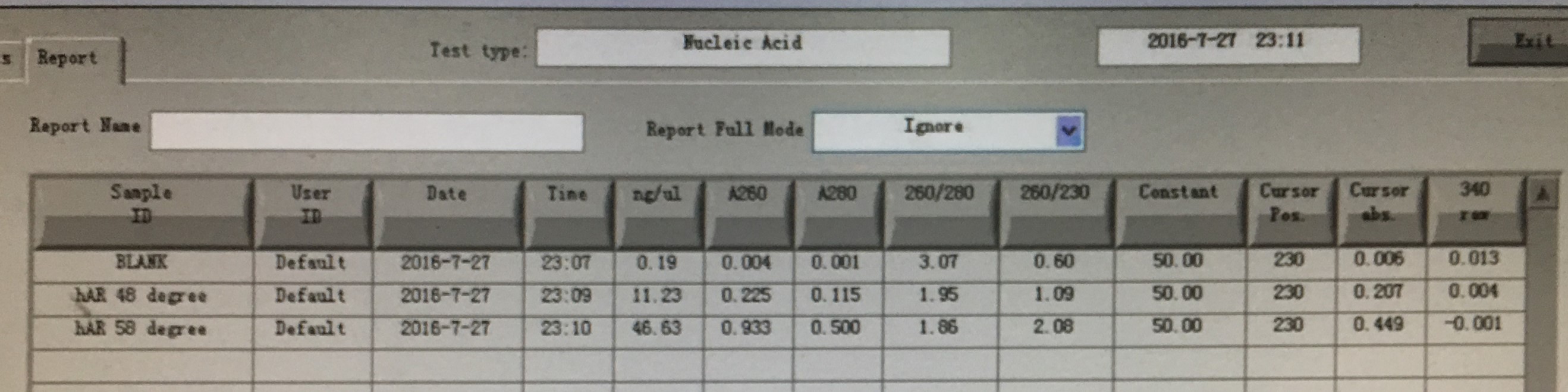
- hAR gene fragment PCR(ADH )
PCR system: 50 μl x 3 tubes
| Name | volume(μl) |
| template | GAL-hAR | 0.5 |
| primer | TRP-ADH-Har | 1 |
PCR conditions:
| Name | Temperature | Duration |
| Pre-denaturation | 95℃ | 3 min |
| Denaturation | 95℃ | 40s | 39x |
| Anealing | 48℃ | 30s | |
| Extension | 68℃ | 1min 30s | |
| Final extension | 72℃ | 10min |
electrophoresis:
| Gel | PCR product | 6 x Loading buffer | DL2000 |
| 1% agarose | 5μl | 1μl | 5μl |
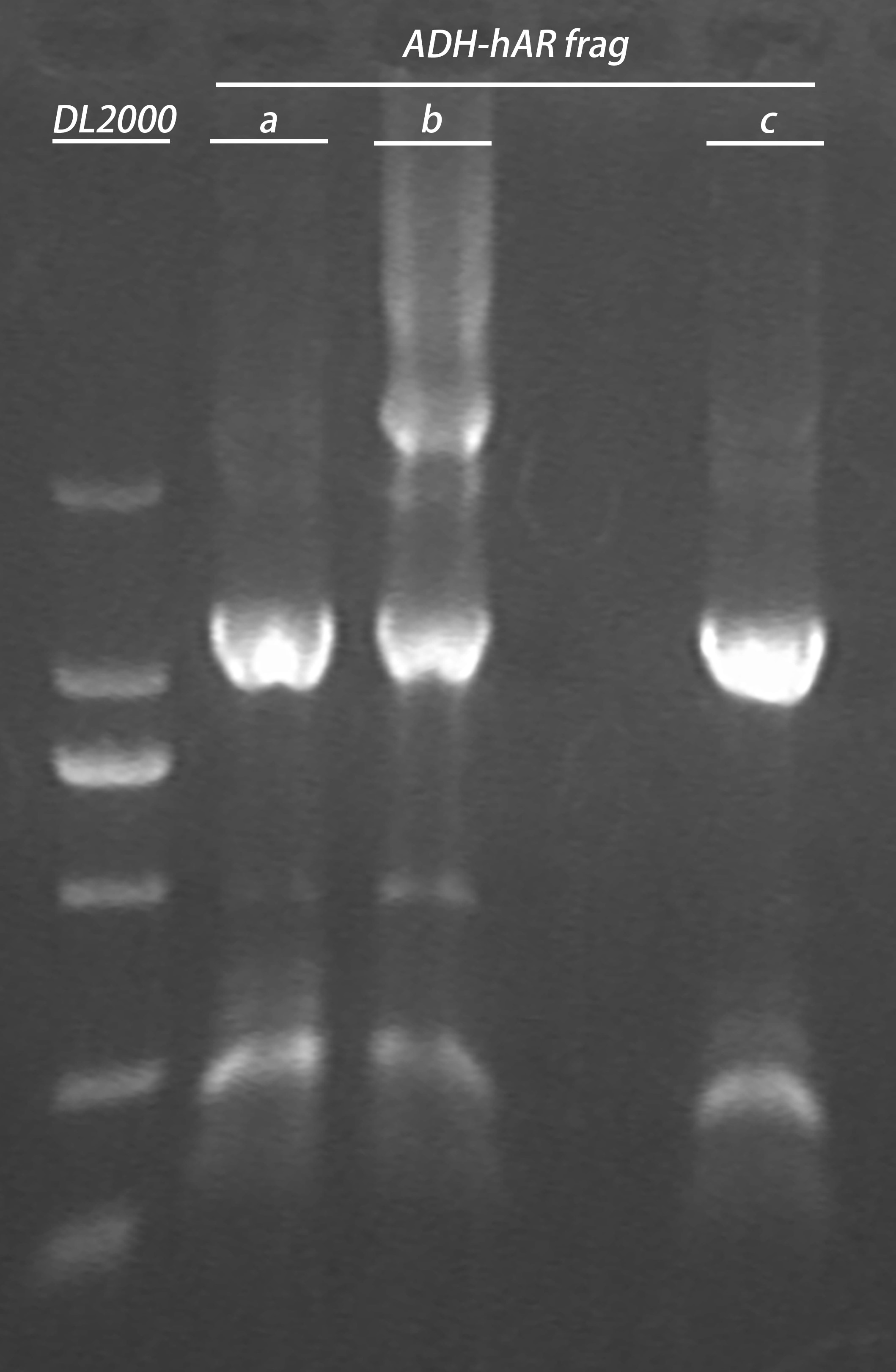
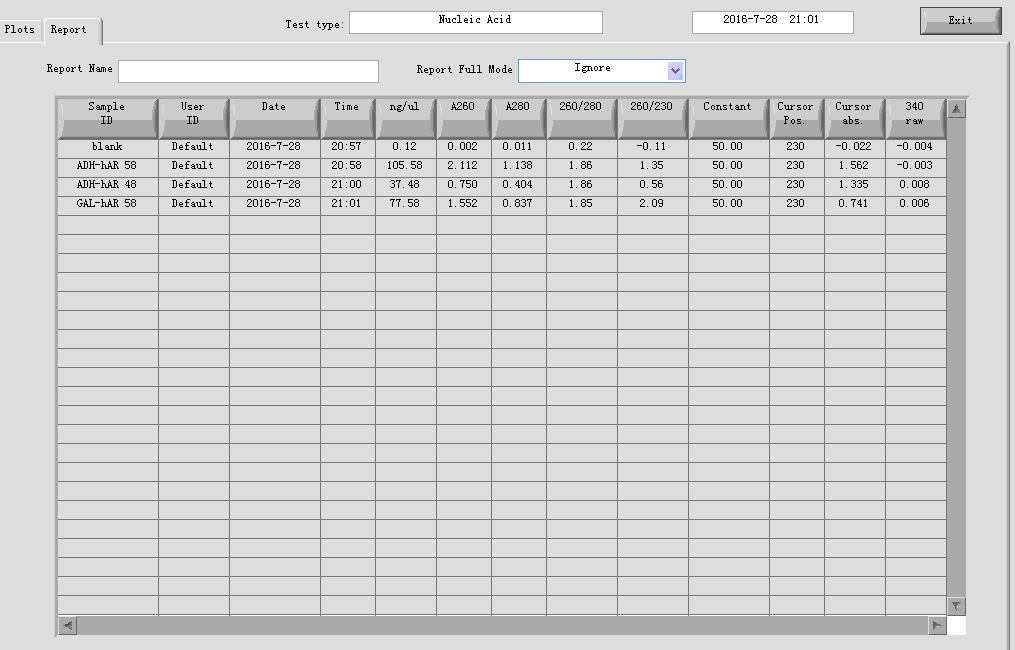
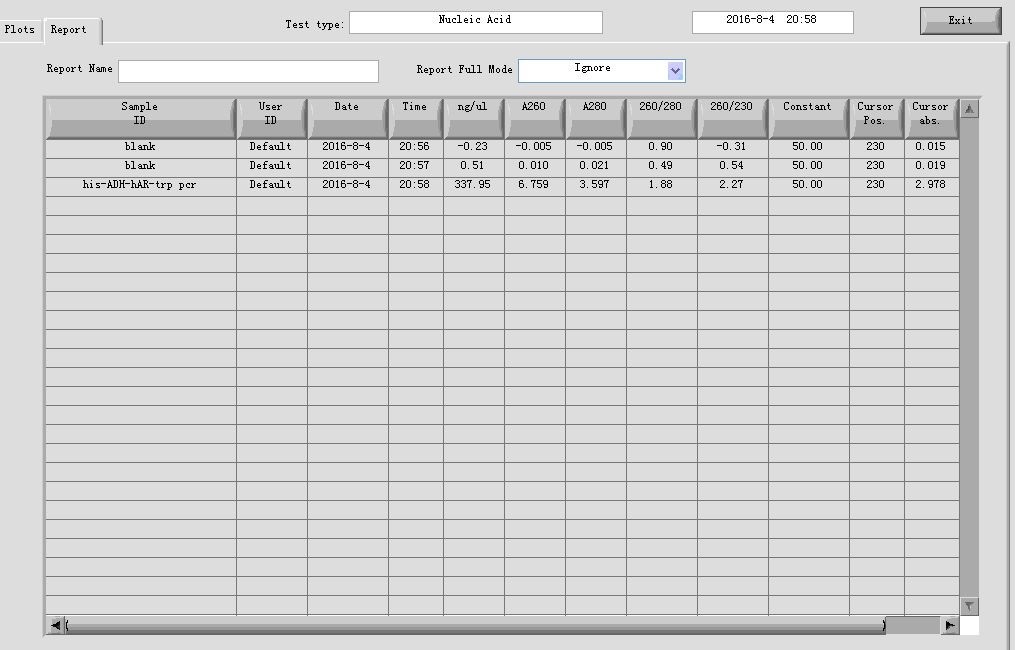
- PCR cleaning
Elution volume: 30μl;3 in 1 tube
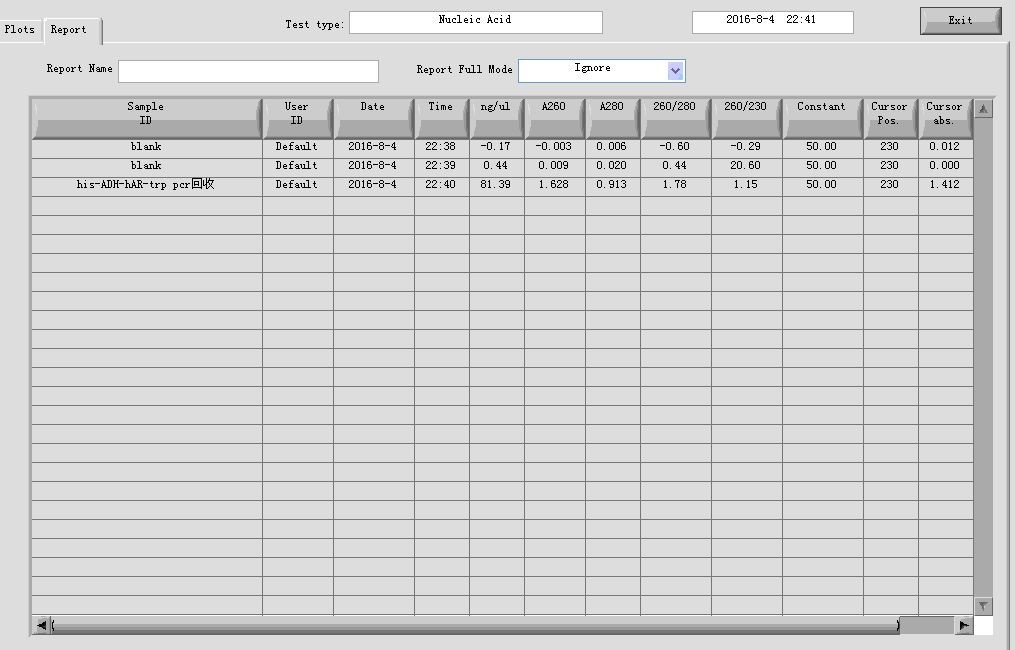
- Gel recovery
electrophoresis:
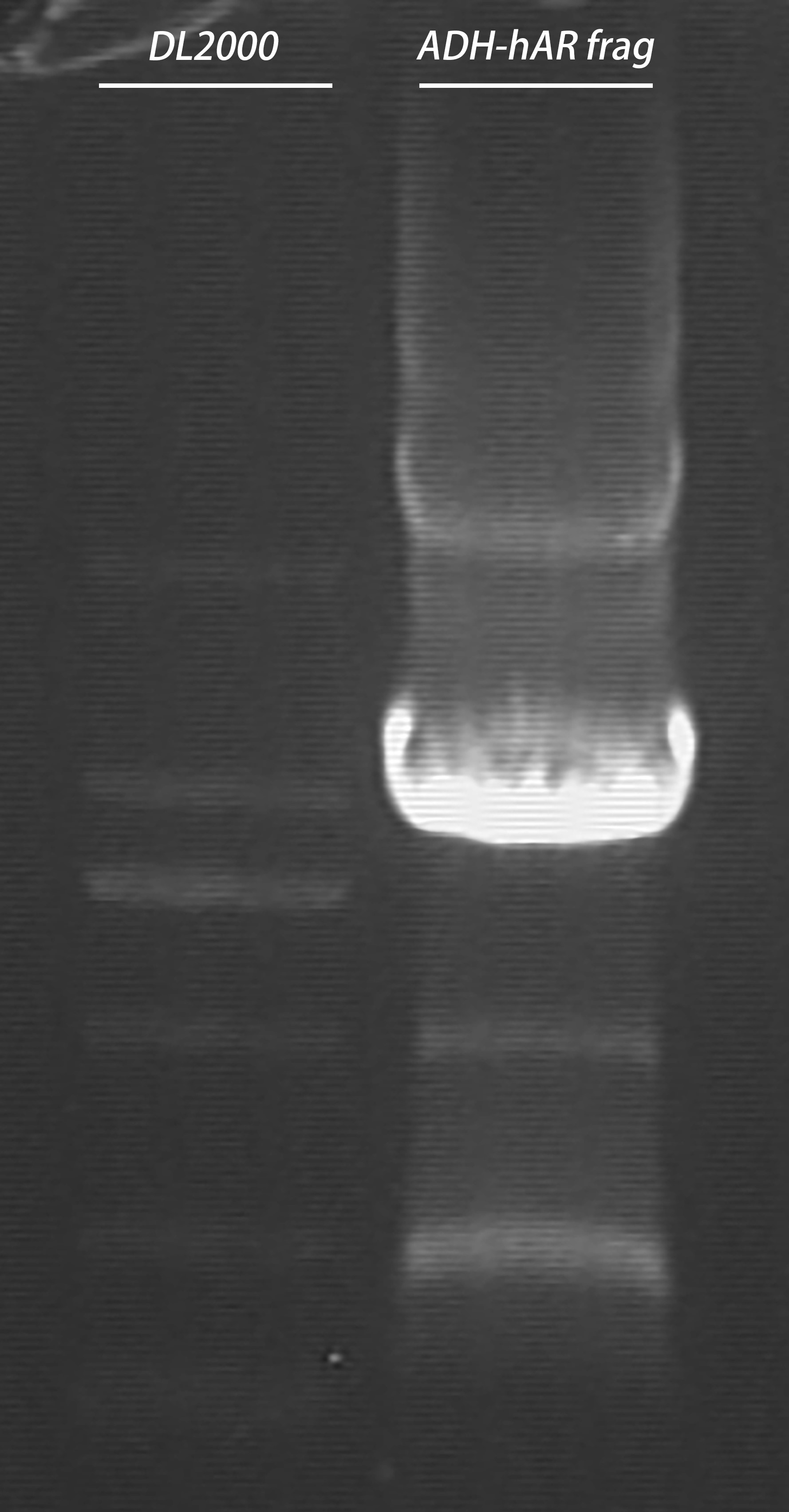
| Gel | PCR cleaning product | 6 x Loading buffer | DL2000 |
| 1% agarose | 30μl | 6μl | 10μl |
gel recovery:
20160805
- Ligation using recombinase
Ligation process were done followed the standardized protocol from the manufacturer, the plasmids and DNA volume were set as follows:
| Construct | Plasmid | DNA | Total volume |
| TRP-ADH-hAR | 1.6 μl |
| TRP-ADH(切) | 0.7 μl |
| ADH-hAR | 20 μl |
37℃ 0.5h
- Transformation of DH5a competent cells
| construct name | DNA volume | Competent cells | LB media | Spread on plate |
| TRP-ADH-hAR | 10 μl | 50 μl | 600 μl | 300 μl |
| GAL-3HA-UG27 | 6 μl | 50 μl | 600 μl | 300 μl |
20160806
- Pick up colonies
TRP-ADH-hAR: 5
GAL-3HA-UG27: 2
- Colony PCR of construct TRP-ADH-hAR
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Culture | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:
TRP-ADH-hAR-R/ TRP-ADH-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 30x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
- Gel electrophoresis of PCR products in Step.1
| Gel | PCR product | 6 x Loading buffer | Volume loaded | DNA ladder |
| 1% agarose | 20μl | 4μl | 10 μl | 5μl |
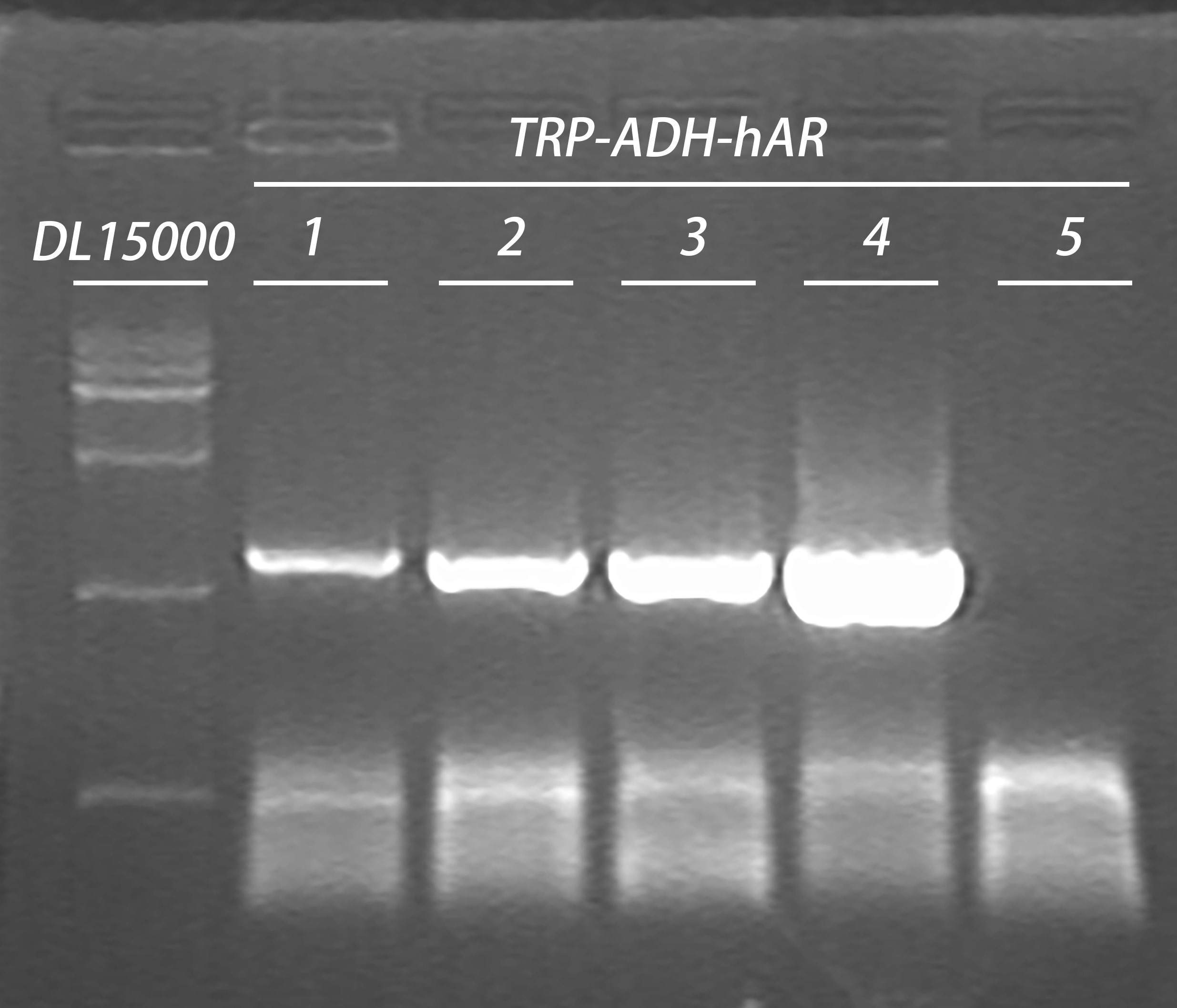
20160807
- Plasmid extraction
TRP-ADH-hAR 1,2,3,4
Elution: 40μl in ddH2O
- Endonulease digestion
37℃, 4.5h
| Plasmid |
| (TRP-ADH-hAR 1/LD-BS-HIS 3) | 12μl |
| Endonuclease | 1μl HindIII (HF) |
| 1μl ApaI |
| Buffer | 5μl Cutsmart |
| ddH2O | 31μl |
Gel electrophoresis
| Gel | Digestion product | 6 x Loading buffer | DL15000 |
| 1% agarose | 50μl | 10μl | 10μl |
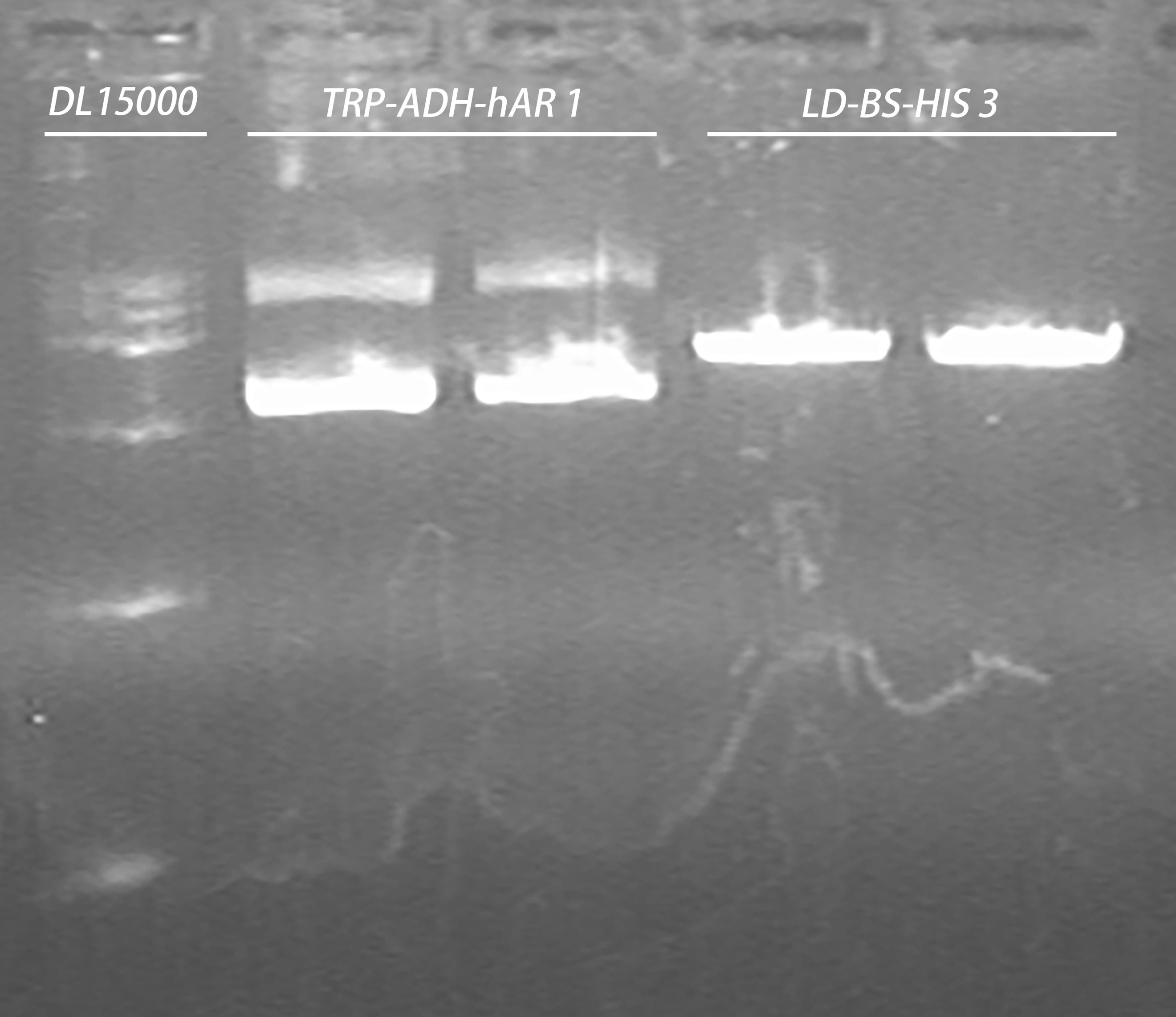
- Gel recovery
Elution: 40μl in ddH2O
- Ligation using T4 ligase
Ligation process were done followed the standardized protocol from the manufacturer, the plasmids and DNA volume were set as follows:
| Construct | Plasmid | DNA | Total volume |
| HIS-ADH-hAR | 6.7 μl LD-BS-HIS3 | 13 μl TRP-ADH-hAR | 20 μl |
22℃ 1h
- Transformation of DH5a competent cells
| construct name | DNA volume | Competent cells | LB media | Spread on plate |
| HIS-ADH-hAR | 10 μl | 50 μl | 600 μl | 300 μl |
20160808
- Pick up colonies
HIS-ADH-hAR: 4 (1pm) + 2 (7:30pm)
- Colony PCR of construct TRP-ADH-hAR (4 picked up at 1 pm )
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Culture | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:
TRP-ADH-hAR-R/ TRP-ADH-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 30x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
Gel electrophoresis of PCR products:
| Gel | PCR product | 6 x Loading buffer | Volume loaded | DNA ladder |
| 1% agarose | 20μl | 4μl | 10 μl | 5μl |
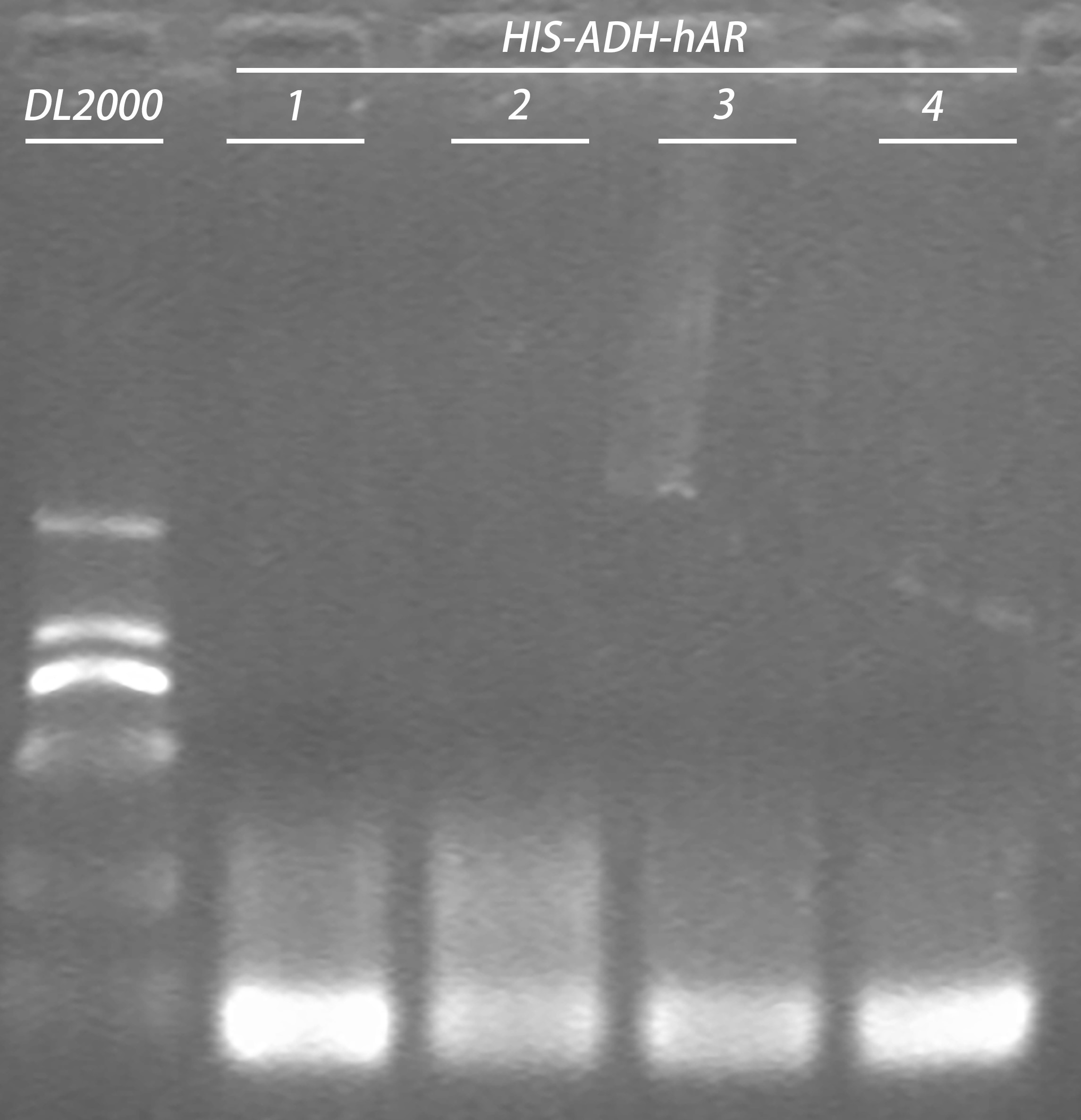
20160809
- Colony PCR of construct TRP-ADH-hAR (3 picked up at 1 pm, 1 picked up at 7:30 pm )
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Culture | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:
TRP-ADH-hAR-R/ TRP-ADH-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 30x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
Gel electrophoresis of PCR products:
| Gel | PCR product | 6 x Loading buffer | Volume loaded | DNA ladder |
| 1% agarose | 20μl | 4μl | 10 μl | 5μl |
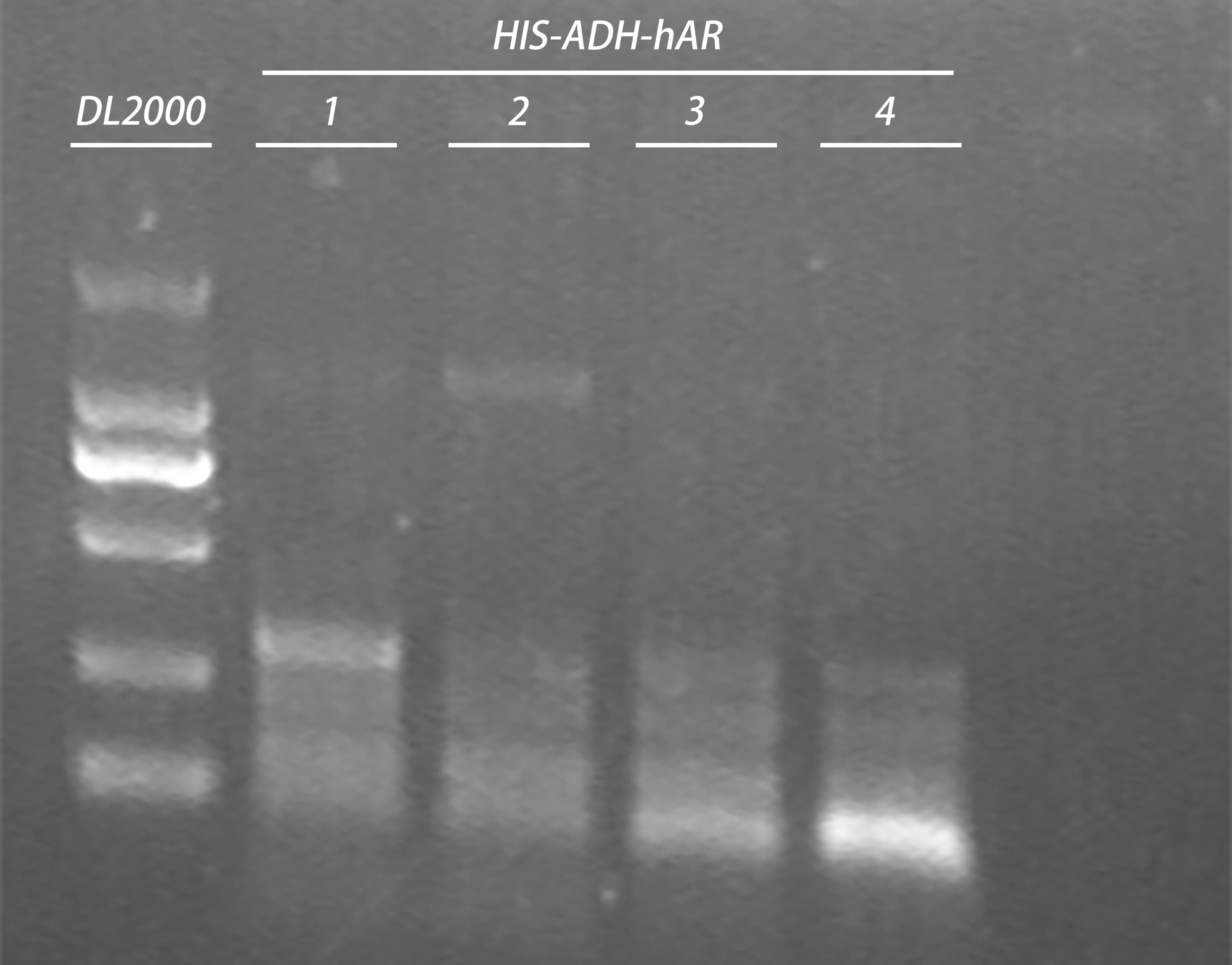
- Plasmid extraction
HIS-ADH-hAR: 2,3
Elution: 40μl in ddH2O
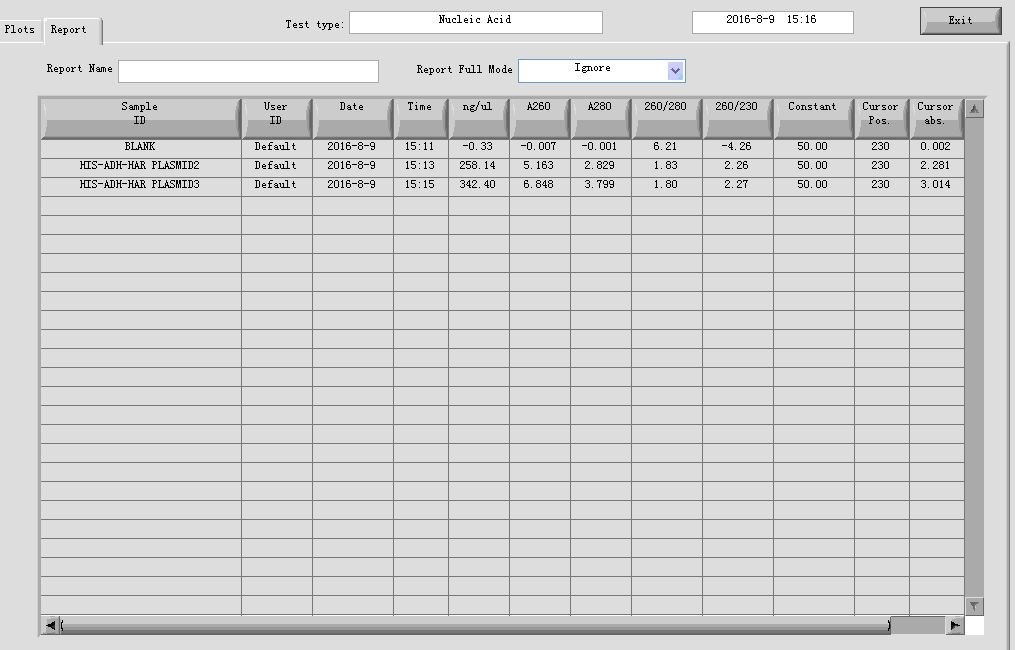
- Taq PCR of plasmid
20 μl system:
| Reagents | Volume |
| Taq | 1 μl |
| Buffer | 2 μl |
| 5μM dNTPs | 1.76 μl |
| Extracted plasmid | 1 μl |
| 10 pmol/µl Primer | 0.4x2 μl |
| ddH2O | 14.04 μl |
Primer:
TRP-ADH-hAR-R/ TRP-ADH-hAR-F
PCR condition was set as follows:
| Name | Temperature | Duration |
| Pre-denaturation | 94℃ | 3 min |
| Denaturation | 94℃ | 40s | 30x |
| Anealing | 60℃ | 30s | |
| Extension | 72℃ | 2min | |
| Final extension | 72℃ | 7min |
Gel electrophoresis of PCR products:
| Gel | PCR product | 6 x Loading buffer | Volume loaded | DNA ladder |
| 1% agarose | 20μl | 4μl | 10 μl | 5μl |
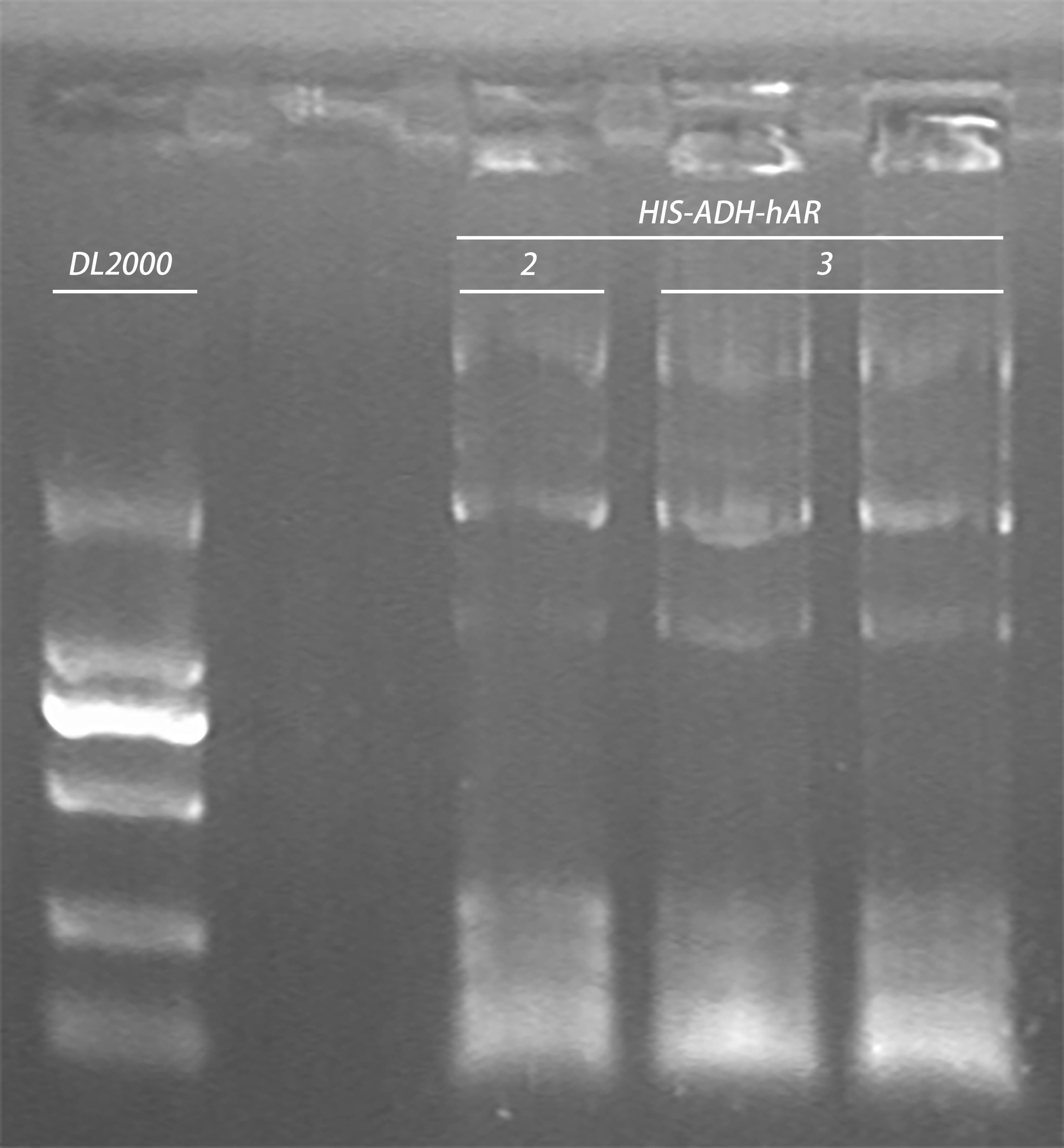
- re-inoculate yeast strain construct (10:15 pm)
| yeast strain | construct | media |
| 9060 | - | 5ml YPD/ YPD agr |
| | ADH-NLc (9060-4) | 5ml YPD |
| | ADH-NLc (ALL) | YPD agar |
| DJ03 | - | 5ml YPD/ YPD agr |
| | ADH-NLc (DJ03-4) | 5ml YPD |
| | ADH-NLc (ALL) | YPD agar |
20160811
- Sequencing
20160812
- Endonuclease digestion
TRP-CU-NLc/ TRP-GAL-NLc digestion is aimed for transformation of yeast cells.
| Plasmid | Enzyme | Buffer | ddH2O |
| TRP-CU-NLc 1 | 4 μl | 0.5 μl NheI-HF |
| 0.5 μl SacI-HF | 5 μl CutSmart | 9 μl |
| TRP-GAL-NLc 1 | 4 μl | 0.5 μl NheI-HF |
| 0.5 μl SacI-HF | 5 μl CutSmart | 9 μl |
| TRP-ADH-hAR 1 | 12 μl | 1 μl HindIII-HF |
| 1 μl ApaI | 5 μl CutSmart | 31 μl |
| LD-BS-HIS | 12 μl | 1 μl HindIII-HF |
| 1 μl ApaI | 5 μl CutSmart | 31 μl |
- KOD PCR for yeast transformation
| Template | Primer |
| HIS-ADH-hAR 3 | BLKS-HIS F/R |
| HIS-GAL-hAR 3b | BLKS-HIS F/R |
| KOD buf. | 5μl |
| dNTPs | 5 μl |
| Mg2+ | 3 μl |
| Primer | 1x2 μl |
| Template | 1 μl |
| KOD pol | 1 μl |
| ddH2O | 34 μl |
| 总 | 50 μl |
- Put yeast colonies into liquid media
| Name | media |
| 9060-1 | 10 ml YPD |
| 9060-TRP-ADH-NLc-1 | 5 ml YPD |
| 9060-TRP-ADH-NLc-2 | 5 ml YPD |
| 9060-TRP-ADH-NLc-4 | 10 ml YPD |
| DJ03-TRP-ADH-NLc-4 | 10 ml YPD |
- Make the construct HIS-ADH-hAR again, all procedures were as before.
Endonuclease digestion: DL15000 5μl
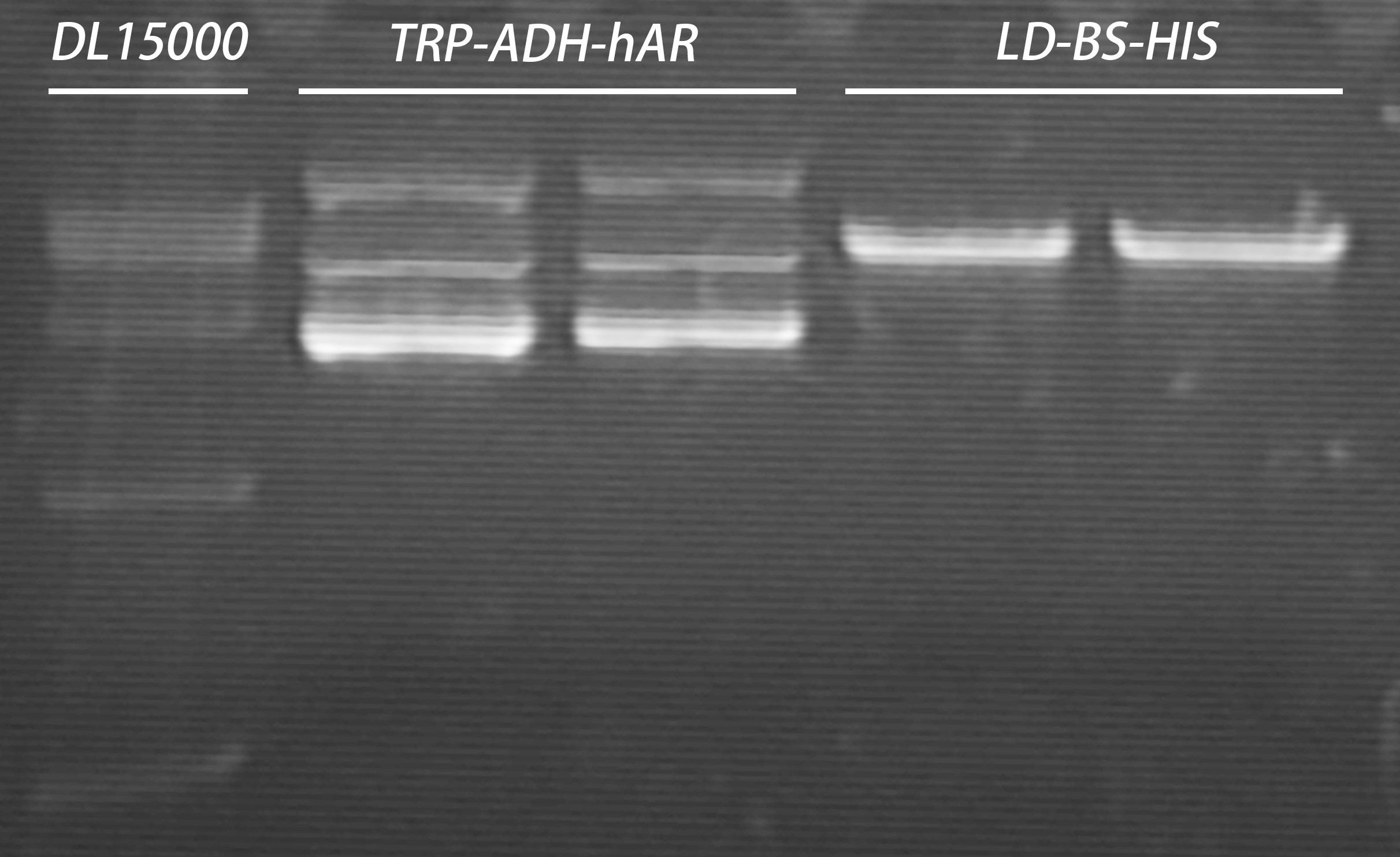 Gel recovery elution volume: 30 μl
Gel recovery elution volume: 30 μl
 Transformation of DH5a cells: 50 μl cells left after last time usage,and last time’s plate
Transformation of DH5a cells: 50 μl cells left after last time usage,and last time’s plate
20160813
- Transformation of yeast cells
| Name | Construct | Constuct volume | plate |
| 9060-1 | TRP-CU-NLc3/ TRP-GAL-NLc3 | 20 μl | TRP- |
| 9060-TRP-ADH-NLc-1 | HIS-GAL-hAR | 20 μl | HIS- |
| 9060-TRP-ADH-NLc-2 | HIS-ADH-hAR | 20 μl | HIS- |
| 9060-TRP-ADH-NLc-4 | HIS-GAL-hAR/ HIS-ADH-hAR | 20 μl | HIS- |
| DJ03-TRP-ADH-NLc-4 | HIS-GAL-hAR/ HIS-ADH-hAR | 10 μl | HIS- |
- Pick up colonies of construct HIS-ADH-hAR as before
20160814
- Colony PCR for construct HIS-ADH-hAR
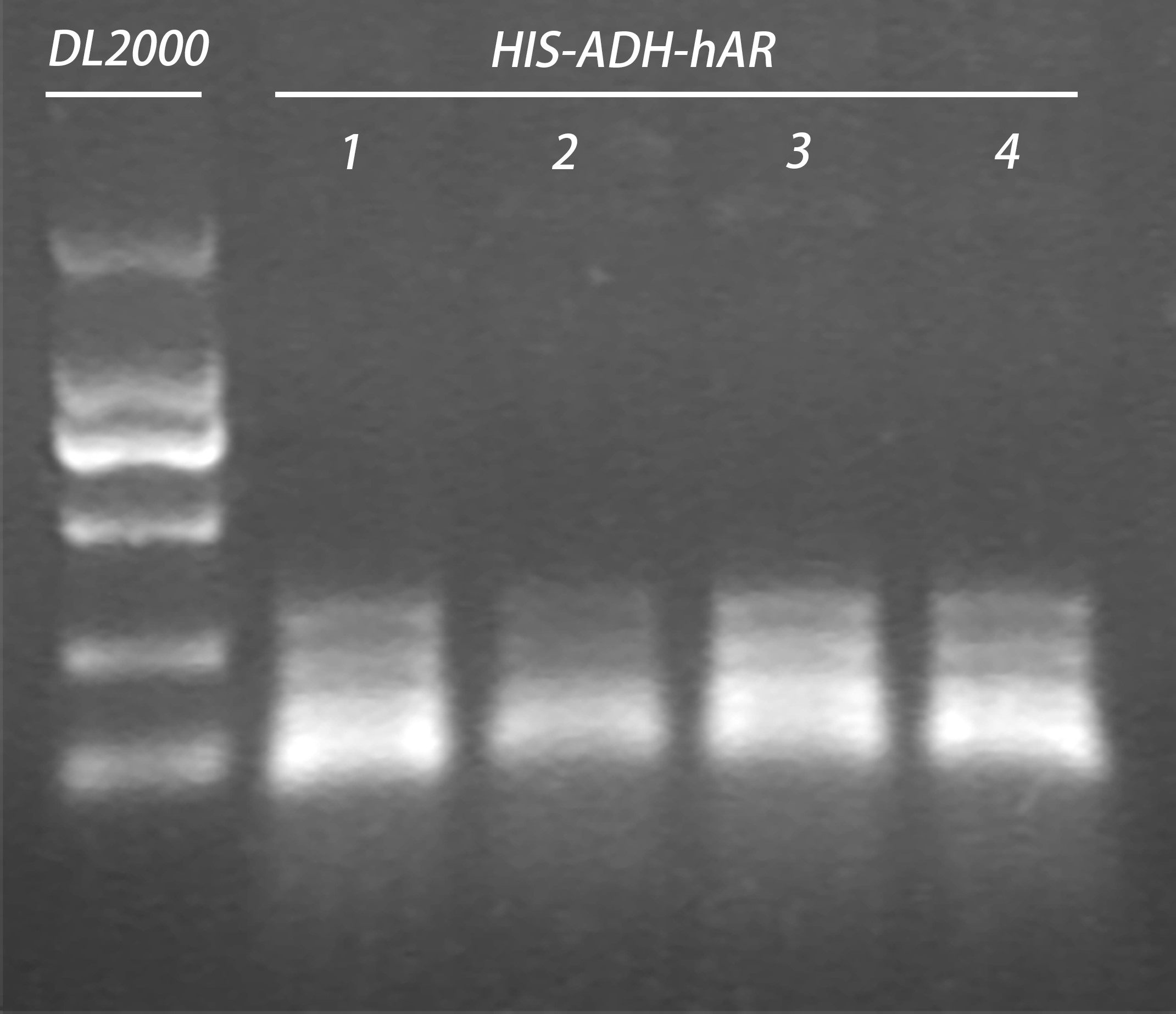
- Plasmid extraction of construct HIS-ADH-hAR
- Pick up anthor 10 colonies from plate of construct HIS-ADH-hAR
20160815
- Colony PCR of 10 colonies from yesterday
- Enzyme digestion of the plasmid extracted yesterday
HindIII-HF
ApaI
CutSmart
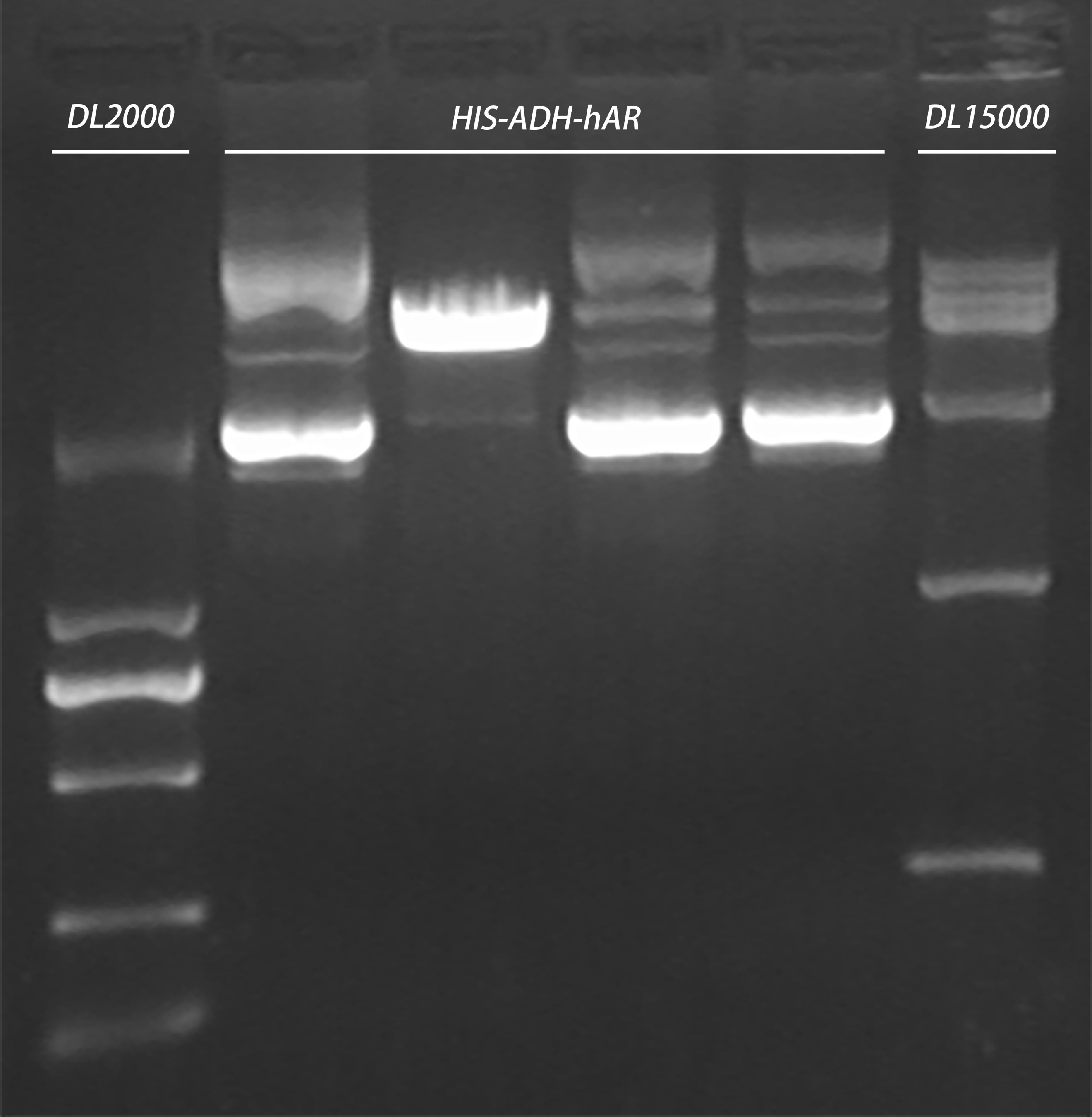
20160816
- The coelenterazine was put into the liquid nitrogen storage pot, MG group, position 6-1 (Red tube lid).
- Pick 8 colonies from TRP-CU-NLc and TRP-GAL-NLc plate and inoculate them on the TRP- plate.
- Taq PCR
| Primer | Template |
| BLKS-HIS R/F | HIS-ADH-hAR 1,2,3,4 |
| HIS-ADH-hAR 3,2 |
| LD-BS-HIS |
| HIS-GAL-Har 3b |
| TRP-ADH-hAR R/F | HIS-ADH-hAR 1,2,3,4 |
| HIS-ADH-hAR 3,2 |
| TRP-ADH-hAR 1 |
| TRP-GAL-hAR R/F | HIS-GAL-Har 3b |
| TRP-GAL-Har 1 |
| HIS-GAL-Har 2b |
- Endonuclease digestion
| Template | Endonuclease | Buffer |
| HIS-ADH-hAR 2,4 | EcoR I | Green FD |
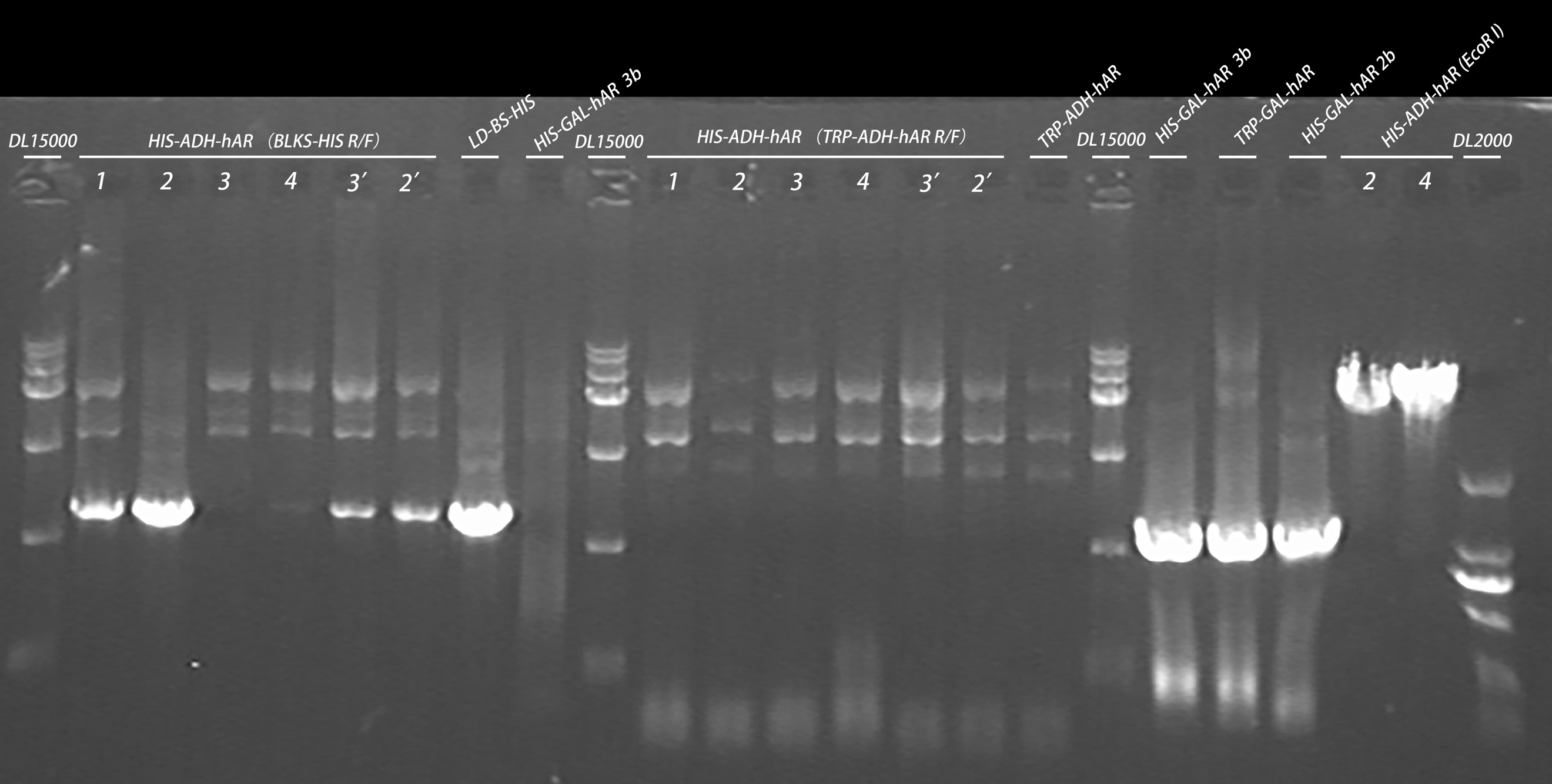
20160820
- Test of the samples using KOD pcr
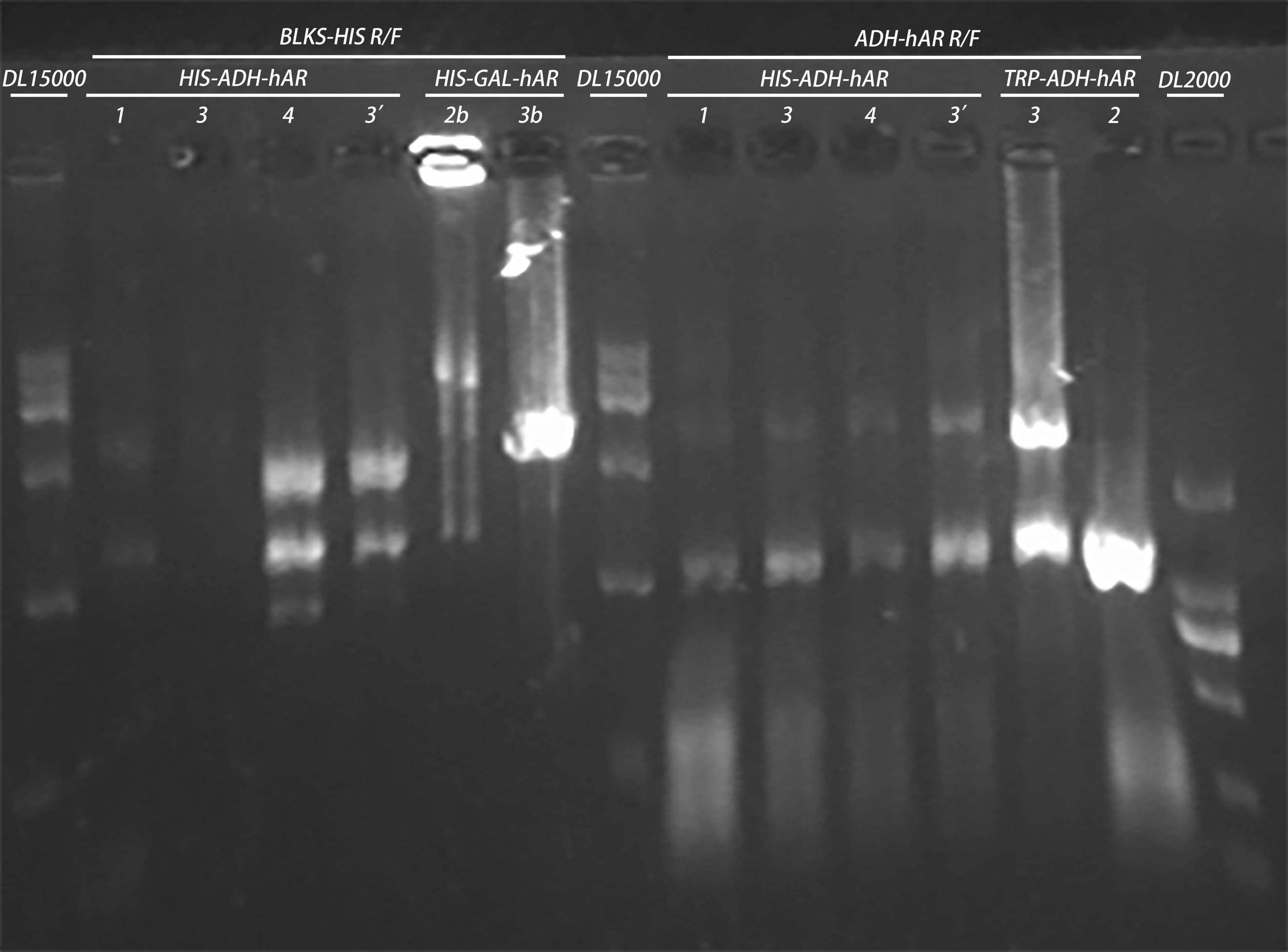
20160827
- Transformation of ADH-NLc1/GAL-NLc1/CU-NLc1 yeast construct
His-GAL-Har 2b/2c/3b
Endonuclease digestion
| Template | Endonuclease | Buffer |
| His-GAL-Har 2b/2c/3b | AflII/ NcoI | 3.1 |
- TRP-ADH plasmid
| Template | Endonuclease | Buffer | Template V |
| TRP-ADH plasmid | ClaI/AscI | CutSmart | 20μl |
- KOD PCR of hAR
| Template | primer | Tm |
| hG/A549 | ADH-hAR-F/R | 58 |
- Ligation using recombinase
ADH-hAR (not cleaned or gel recovered) + TRP-ADH (previous)
- Transformation of DH5a
20160828
- Glucose sensing detection using plate reader
ADH GAL ADH/GAL/9060-1 9060-1
0
5
25
50
100
200
- Characterizing of GAL/CU/9060-1 under flo
20160829
- Enzyme digestion and gel recovery of TRP-ADH-hAR
- Ligation using T4 DNA ligase
TRP-ADH-hAR (gel recovered) + HIS (previous)
20160831
- Glucose sensing detection using plate reader
36h in YPGal
ADH GAL CU 9060-1
0
5
25
50
100
200
- Colony PCR of HIS-ADH-hAR/TRP-ADH-hAR
- Freeze-dring of yest strain ADH-NLc/GAL-NLc/CU-Nlc/9060-1
- Revitalization of freeze-dried yeast strain CU-NLc
3h in culture + cryoprotectant -> 100μl in 5ml YPD
20160901
- Plasmid extraction of TRP-ADH-hAR/HIS-ADH-hAR
- Endonuclease digestion of HIS-ADH-hAR 1/2/3 and transformation of yeast strain TRP-ADH-NLc
- Freeze drying of yeast strain TRP-ADH-NLc and 9060-1
20160902
- Shake the yeast strain TRP-ADH-NLc1/TRP-GAL-NLc2/TRP-CU-NLc4/9060-1 for freeze drying and reactivation test
20160904
- KOD PCR of ADH-NLc 1 / GAL-NLc 2 / CU-NLc 4 / 9060 1
| KOD buf. | 5μl |
| dNTPs | 5 μl |
| Mg2+ | 3 μl |
| Primer | 1x2 μl |
| Template | 1 μl |
| KOD pol | 1 μl |
| ddH2O | 33 μl |
| Total | 50 μl |
| Name | Temperature | Duration |
| Pre-denaturation | 95℃ | 5 min |
| Denaturation | 95℃ | 40s | 39x |
| Anealing | 58℃ | 30s | |
| Extension | 68℃ | 2min | |
| Final extension | 72℃ | 10min |
| Hold | 16℃ | infinite |
|
| Number | Primer | Template (each 3 tubes) |
| 1 | CU-hAR-R/CU-hAR-F2 | TRP-ADH-hAR | TRP-ADH-hAR |
| 2 | GAL-hAR-Rhis/GAL-hAR-F | TRP-ADH-hAR | hG DNA |
| 3 | GAL-hAR-R/GAL-hAR-F | TRP-ADH-hAR | hG DNA |
| 4 | ADH-hAR-Rhis/ADH-hAR-Fhis | TRP-ADH-hAR | hG DNA |
- 2 displays no result
- Ligation using recombinase and transformation of DH5a cells
| Vector | DNA fragment |
| TRP-GAL (hAR cut) | hAR (GAL) |
| TRP-ADH (hARcut) | hAR-his (ADH) |
- Colony PCR of transformed yeast cells (HIS-ADH-hAR)
primer: ADH-hAR-R/ADH-hAR-F
- Inoculate 4 colonies (ADH-NLc/ GAL-NLc/ CU-NLc/ 9060) in YPGal media
20160905
- Inoculate 4 colonies (ADH-hAR1/2/3/4) in YPD media
- Colony PCR of the construct TRP-GAL-hAR/TRP-ADH-hAR-his (failed, used DL15000 as ladder )
20160906
- Epinephrine response measurement
Culture process: 16 h in YPD -> 1 h in YPGal
| EP (μM) | ADH-hARa | ADH-hARb | ADH-hARc | ADH-hARd | ADH-NLc | 9060 |
| 0 | | | | | | | | | | | | |
| 10 | | | | | | | | | | | | |
| 25 | | | | | | | | | | | | |
| 50 | | | | | | | | | | | | |
| 75 | | | | | | | | | | | | |
| 100 | | | | | | | | | | | | |
| 500 | | | | | | | | | | | | |
| 1000 | | | | | | | | | | | | |
200 μl total volume, coelenterazine h in 10 μM final concentration
- Colony PCR of the construct TRP-GAL-hAR/TRP-ADH-hAR-his, extract the plasmids
20160907~20160911
- Construction of yeast genomic integration plasmid HIS-ADH-hAR-his, HIS-GAL-hAR-his and HIS-ADH-Gpa2M
20160912~20160918
- Transformation of yeast cells with HIS-ADH-hAR-his, HIS-GAL-hAR-his and HIS-ADH-Gpa2M and measure the response ability in glucose or epinephrine receptor or mimic pathological solutions.
- Test of the response ability after freeze drying process.
20160912~20160918
- Making our parts in pSB1C3 to finish the final step!
20160919~20161023
- Synthesized DNA from GENEWIZ to make further modification of ADRB2
- Dealing with the wiki work and presentation draft.
- Western blotting of His-tagged proteins and Immunocytochemistry analysis of membrane localization.
20161023~20161025
- Get good sleep to compensate for the coming jet-lag.
20161026
- Set off for Giant-Jamboree!



 Gel recovery elution volume: 30 μl
Gel recovery elution volume: 30 μl
 Transformation of DH5a cells: 50 μl cells left after last time usage,and last time’s plate
Transformation of DH5a cells: 50 μl cells left after last time usage,and last time’s plate














































