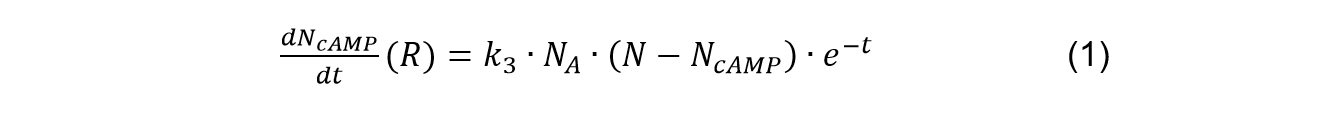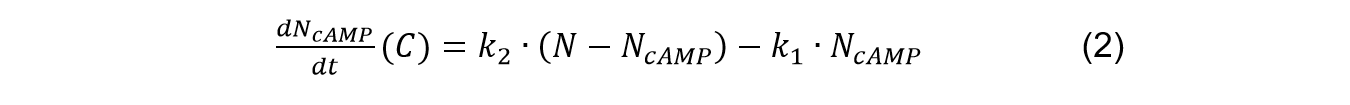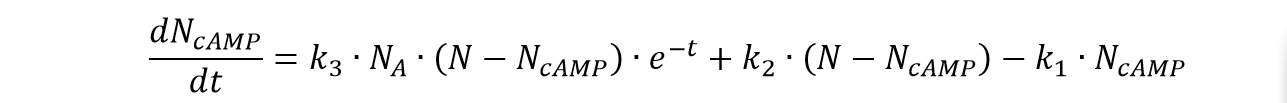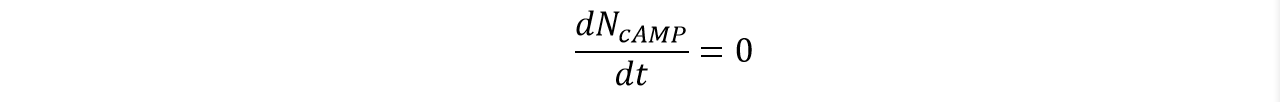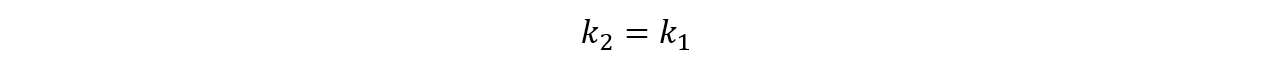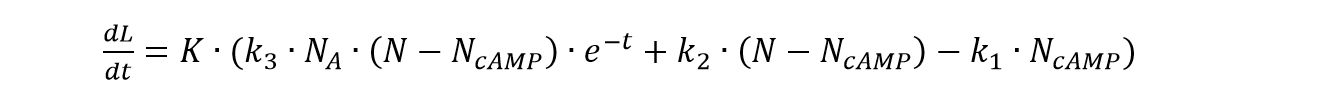|
|
| (20 intermediate revisions by 3 users not shown) |
| Line 1: |
Line 1: |
| | + | {{SJTU-BioX-Shanghai/Navigator}} |
| | <html> | | <html> |
| − | <div id="navigator">
| |
| − | <aside id="nav">
| |
| − | <p><a href="/File:T--SJTU-BioX-Shanghai--logov2.jpg" class="image" title="float = left"><img alt="float = left" src="/wiki/images/8/8f/T--SJTU-BioX-Shanghai--logov2.jpg" width="250" height="120"/></a>
| |
| − | </p>
| |
| − |
| |
| − | <nav>
| |
| − | <ul id="nav2">
| |
| − | <li id="home"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai">home</a></li>
| |
| − | <li id="project">
| |
| − | <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Description">Project</a>
| |
| − | <ul>
| |
| − | <li><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Description">Description</a></li>
| |
| − | <li><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Motivation">Motivation</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Design"> Design as a whole </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Yeast_glucose_sensor"> Yeast glucose sensor </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Yeast_epinephrine_sensor"> Yeast epinephrine sensor </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Yeast_biosensor_as_freeze-dried_powders"> Yeast biosensor as freeze-dried powders </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Vision_for_the_future">Vision for the future</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Experiments"> Protocols </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Notebook"> Notebook </a></li>
| |
| − | </ul>
| |
| − | </li>
| |
| − |
| |
| − | <li id="safety"> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Safety"> Safety </a> </li>
| |
| − | <li id="parts"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Parts">Parts</a>
| |
| − | <ul>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Parts">Team Parts </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Composite_Part"> Composite Parts </a></li>
| |
| − | </ul>
| |
| − | </li>
| |
| − |
| |
| − | <li id="model"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Model">Model</a>
| |
| − | <ul>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Yeast_activation"> Yeast activation </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Yeast_luminescence_process"> Yeast luminescence process </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Combination_and_use"> Combination and use </a></li>
| |
| − | </ul>
| |
| − | </li>
| |
| − |
| |
| − | <li id="hp"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices"> Human Practice </a>
| |
| − | <ul>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices">Introduction </a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices/Bioethics">Bioethics</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices/Hospital_Experiencing">Hospital Experiencing</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices/Marketing">Marketing</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices/Medical_Service">Medical Service</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Human_Practices/Outreach">Outreach</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Integrated_Practices"> Integrated Practices </a></li>
| |
| − | </ul>
| |
| − | </li>
| |
| − | <li id="achievement"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Achievement"> Achievement </a>
| |
| − | <ul>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Achievement">Achievement</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/HP/Gold">Human Practice:Gold and Silver </a></li>
| |
| − | </ul>
| |
| − | </li>
| |
| − |
| |
| − | <li id="team"><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Team">Team </a>
| |
| − | <ul>
| |
| − | <li><a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Team">Team</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Collaborations">Collaborations</a></li>
| |
| − | <li> <a href="https://2016.igem.org/Team:SJTU-BioX-Shanghai/Attributions">Attributions</a></li>
| |
| − |
| |
| − | </ul>
| |
| − | </li>
| |
| − | </ul>
| |
| − | </nav>
| |
| − | </div>
| |
| − | <style>
| |
| − | body {
| |
| − | behavior:url(csshover.htc);
| |
| − | }
| |
| − | #nav{
| |
| − | position: fixed;
| |
| − | left:0px;
| |
| − | top:10px;
| |
| − | padding-top: 0px;
| |
| − | padding-left: 0px;
| |
| − | width:100%;
| |
| − | height:135px;
| |
| − | background-color:#f4b045;
| |
| − | }
| |
| − | ul#nav2 {
| |
| − | position: absolute;
| |
| − | right:0px;
| |
| − | top:5px;
| |
| − | padding:0px;
| |
| − | margin:0px;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li {
| |
| − | float:left;
| |
| − | width:120px;
| |
| − | height:120px;
| |
| − | list-style:none;
| |
| − | background-color:#f4b045;
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li#home {
| |
| − | background-image:url(/wiki/images/4/4d/T--SJTU-BioX-Shanghai--home.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#project {
| |
| − | background-image:url(/wiki/images/4/4e/T--SJTU-BioX-Shanghai--project.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#safety {
| |
| − | background-image:url(/wiki/images/1/19/T--SJTU-BioX-Shanghai--safety.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#parts {
| |
| − | background-image:url(/wiki/images/c/ca/T--SJTU-BioX-Shanghai--parts.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#model {
| |
| − | background-image:url(/wiki/images/d/de/T--SJTU-BioX-Shanghai--model.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#hp {
| |
| − | background-image:url(/wiki/images/b/bb/T--SJTU-BioX-Shanghai--human_practice.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#achievement {
| |
| − | background-image:url(/wiki/images/d/d2/T--SJTU-BioX-Shanghai--achievements.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li#team {
| |
| − | background-image:url(/wiki/images/f/f1/T--SJTU-BioX-Shanghai--team.jpg);
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 120px;
| |
| − | }
| |
| − | ul#nav2 li li {
| |
| − | float:left;
| |
| − | width:120px;
| |
| − | height:35px;
| |
| − | list-style:none;
| |
| − | background-color:#f4b045;
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 35px;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li ul {
| |
| − | display:none;
| |
| − | margin:0;
| |
| − | padding:0;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li:hover ul{
| |
| − | display:block;
| |
| − | background-color:#ff9600;
| |
| − | background-repeat:no-repeat;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li li:hover {
| |
| − | background-color:#ff9600;
| |
| − | background-repeat:no-repeat;
| |
| − | background-size:120px 35px;
| |
| − | z-index=10;
| |
| − | }
| |
| − |
| |
| − | ul#nav2 li a{
| |
| − | font: 16px/20px STXihei, sans-serif !important;
| |
| − | display:block;
| |
| − | border:0px;
| |
| − | text-decoration:none;
| |
| − | color:#000;
| |
| − | text-align:center;
| |
| − | margin-top:94px;
| |
| − | z-index=10;
| |
| − | }
| |
| − | ul#nav2 li li a {
| |
| − | display:block;
| |
| − | font: 10px/15px STXihei, sans-serif !important;
| |
| − | border:0px;
| |
| − | padding:3px;
| |
| − | text-decoration:none;
| |
| − | text-align:center;
| |
| − | color:#000;
| |
| − | width:120px;
| |
| − | margin-top:0px;
| |
| − | z-index=10;
| |
| − | }
| |
| − |
| |
| − | ul#nav2>li li a {
| |
| − | width:auto;
| |
| − | z-index=10;
| |
| − | }
| |
| − | </style>
| |
| − |
| |
| − | </aside>
| |
| − |
| |
| − | <style>
| |
| − | #toc {display: none;}
| |
| − | </style>
| |
| − |
| |
| − |
| |
| | </p><p> | | </p><p> |
| | <article id="main"> | | <article id="main"> |
| Line 214: |
Line 10: |
| | <br> | | <br> |
| | <h1><span class="mw-headline">Yeast luminescence process</span></h1> | | <h1><span class="mw-headline">Yeast luminescence process</span></h1> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| Line 221: |
Line 16: |
| | <p class="main-page">In this section, we established a model for the quantity relationship between the concentration of adrenalin and the luminance magnitude of yeasts which includes the physiological process as receptor signaling, cAMP-AMP-ADP-ATP-cAMP circulation and reporter protein lightening. After the parameters in the model were adjusted, we found that our simulation results fits experiment results well. | | <p class="main-page">In this section, we established a model for the quantity relationship between the concentration of adrenalin and the luminance magnitude of yeasts which includes the physiological process as receptor signaling, cAMP-AMP-ADP-ATP-cAMP circulation and reporter protein lightening. After the parameters in the model were adjusted, we found that our simulation results fits experiment results well. |
| | </p> | | </p> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | <h2><span class="mw-headline">Basic assumption</span></h2> | | <h2><span class="mw-headline">Basic assumption</span></h2> |
| − | <ol><li>the Adrenalin is uniformly distributed amid yeasts</li> | + | <ol><li>The Adrenalin is uniformly distributed amid yeasts</li> |
| − | <li>the number of yeasts during lightening period is constant and the physical state of each yeasts is the same</li> | + | <li>The number of yeasts during lightening period is constant and the physical state of each yeasts is the same</li> |
| − | <li>the function of the receptor, Gs protein, Adenylate cyclase and reporter protein is not saturated</li> | + | <li>The function of the receptor, Gs protein, Adenylate cyclase and reporter protein is not saturated</li> |
| | <li>During the whole process, the total concentration of cAMP,AMP,ADP,ATP in the cell and the concentration of adrenalin are constant and in the beginning the concentration of cAMP is half of the total concentration of cAMP,AMP,ADP,ATP</li> | | <li>During the whole process, the total concentration of cAMP,AMP,ADP,ATP in the cell and the concentration of adrenalin are constant and in the beginning the concentration of cAMP is half of the total concentration of cAMP,AMP,ADP,ATP</li> |
| | <li>Without stimulation, we assume the concentration of cAMP in cells stay constant</li> | | <li>Without stimulation, we assume the concentration of cAMP in cells stay constant</li> |
| | <li>To simpify the problem,we see the process of the conversion of AMP-ADP-ATP as a whole,which we define Process I.</li> | | <li>To simpify the problem,we see the process of the conversion of AMP-ADP-ATP as a whole,which we define Process I.</li> |
| | </li></ol> | | </li></ol> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | | | |
| | <h2><span class="mw-headline">Variables</span></h2> | | <h2><span class="mw-headline">Variables</span></h2> |
| − | <!--无序列表-->
| |
| | <ul><li>N: the total concentration of cAMP,AMP,ADP,ATP </li> | | <ul><li>N: the total concentration of cAMP,AMP,ADP,ATP </li> |
| − | <li>N<sub>Camp</sub>: the concentration of Camp</li> | + | <li>N<sub>cAMP</sub>: the concentration of cPMP</li> |
| | <li>N<sub>A</sub>:the concentration of Adrenalin</li> | | <li>N<sub>A</sub>:the concentration of Adrenalin</li> |
| − | <li>L:luminance magnitude</li> | + | <li>L:luminance magnitude</li></ul> |
| − | <br> | + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | + | |
| | <h2><span class="mw-headline">Formula and derivation</span></h2> | | <h2><span class="mw-headline">Formula and derivation</span></h2> |
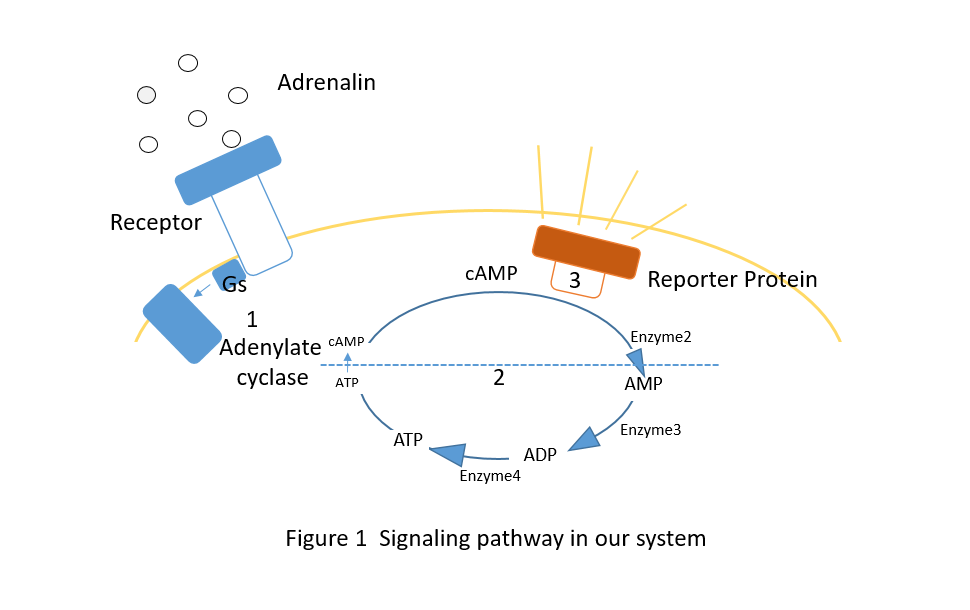
| − | <center><img src="/wiki/images/a/a9/T--SJTU-BioX-Shanghai--figure1-paper2.png" width=60% height=60%/></center> | + | <center><img src="/wiki/images/b/be/T--SJTU-BioX-Shanghai--figure1-paper20.png" width=60% height=60%/></center> |
| | <p class="main-page">As shown in figure1, the signal pathway of Adrenalin-influenced yeasts’ lightening can be divided into three parts: | | <p class="main-page">As shown in figure1, the signal pathway of Adrenalin-influenced yeasts’ lightening can be divided into three parts: |
| | </p> | | </p> |
| Line 255: |
Line 46: |
| | cyclase downstram will be activated and translate ATP to cAMP.</li> | | cyclase downstram will be activated and translate ATP to cAMP.</li> |
| | <li>cAMP-AMP-ADP-ATP-cAMP circulation:to simplify the problem, we see the transformation from AMP to ATP as a whole</li> | | <li>cAMP-AMP-ADP-ATP-cAMP circulation:to simplify the problem, we see the transformation from AMP to ATP as a whole</li> |
| − | <li>Reporter protein lightening: Our receptor protein is only activated by Camp.</li> | + | <li>Reporter protein lightening: Our receptor protein is only activated by cAMP.</li> |
| | </ol> | | </ol> |
| − | <p class="main-page">After analyzing the signal pathway of Adrenalin-influenced yeasts’ lightening, it can be instinctively noticed that the variation of the concentration of Camp is critical in the problem. | + | <p class="main-page">After analyzing the signal pathway of Adrenalin-influenced yeasts’ lightening, it can be instinctively noticed that the variation of the concentration of cAMP is critical in the problem. |
| | </p> | | </p> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | <h3><span class="mw-headline">Receptor signaling</span></h3> | | <h3><span class="mw-headline">Receptor signaling</span></h3> |
| | <ol><li>Based on assumption 3 and the effect of the receptor, we can know that the rate of the formation of cAMP is positively related to the concentration of Adrenalin.</li> | | <ol><li>Based on assumption 3 and the effect of the receptor, we can know that the rate of the formation of cAMP is positively related to the concentration of Adrenalin.</li> |
| − | <li>Besides, since cAMP is formed after Process I, we can know that the rate of the formation of Camp is also positively related to the total concentration of AMP, ADP and ATP in cells.</li> | + | <li>Besides, since cAMP is formed after Process I, we can know that the rate of the formation of cAMP is also positively related to the total concentration of AMP, ADP and ATP in cells.</li> |
| | <li>In reality, the working efficiency of our receptor will decrease during the period of detection. We assume the relationship of its working efficiency corresponding to time as E(t)=e<sup>-t</sup>.</li> | | <li>In reality, the working efficiency of our receptor will decrease during the period of detection. We assume the relationship of its working efficiency corresponding to time as E(t)=e<sup>-t</sup>.</li> |
| | </ol> | | </ol> |
| − | <p class="main-page">Therefore, we can get the situation of the formation of Camp in receptor signaling as: | + | <p class="main-page">Therefore, we can get the situation of the formation of cAMP in receptor signaling as: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/e/e0/T--SJTU-BioX-Shanghai--figure1-paper2a.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/e/e0/T--SJTU-BioX-Shanghai--figure1-paper2a.png" width=60% height=60%/></center> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| − | <h3><span class="mw-headline">Camp-amp circulation</span></h3> | + | <h3><span class="mw-headline">cAMP-AMP-ADP-ATP-cAMP circulation</span></h3> |
| | <p class="main-page">Apart from the formation of cAMP mentioned in the Receptor signaling, there are many other physiological process in cells which can also form cAMP. We see them as a whole and the rate of it is simply positively related to total concentration of AMP, ADP and ATP in cells. | | <p class="main-page">Apart from the formation of cAMP mentioned in the Receptor signaling, there are many other physiological process in cells which can also form cAMP. We see them as a whole and the rate of it is simply positively related to total concentration of AMP, ADP and ATP in cells. |
| | </p> | | </p> |
| | + | |
| | <br> | | <br> |
| − | <br>
| + | <p class="main-page">In the cAMP-AMP-ADP-ATP-cAMP circulation, there are many physiological process in cells which can transform cAMP to AMP and the rate of it is positively related to the concentration of cAMP in cells. |
| − | <p class="main-page">In the Camp-amp circulation, there are many physiological process in cells which can transform cAMP to AMP and the rate of it is positively related to the concentration of cAMP in cells. | + | |
| | </p> | | </p> |
| − | <br>
| + | |
| | <br> | | <br> |
| | <p class="main-page">Therefore, we can get the situation of the formation of cAMP in the circulation as: | | <p class="main-page">Therefore, we can get the situation of the formation of cAMP in the circulation as: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/8/82/T--SJTU-BioX-Shanghai--figure1-paper2b.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/8/82/T--SJTU-BioX-Shanghai--figure1-paper2b.png" width=60% height=60%/></center> |
| | <p class="main-page">Based on equation (1) and (2), we can derive the formulation of the concentration of the cAMP in cells corresponding to the concentration of the Adrenalin as: | | <p class="main-page">Based on equation (1) and (2), we can derive the formulation of the concentration of the cAMP in cells corresponding to the concentration of the Adrenalin as: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/9/9c/T--SJTU-BioX-Shanghai--figure1-paper2c.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/9/9c/T--SJTU-BioX-Shanghai--figure1-paper2c.png" width=60% height=60%/></center> |
| | <p class="main-page">If N<sub>A</sub>=0, then the concentration of cAMP should be constant, that is: | | <p class="main-page">If N<sub>A</sub>=0, then the concentration of cAMP should be constant, that is: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/3/38/T--SJTU-BioX-Shanghai--figure1-paper2d.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/3/38/T--SJTU-BioX-Shanghai--figure1-paper2d.png" width=60% height=60%/></center> |
| | <p class="main-page">Then we can get: | | <p class="main-page">Then we can get: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/8/8f/T--SJTU-BioX-Shanghai--figure1-paper2e.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/8/8f/T--SJTU-BioX-Shanghai--figure1-paper2e.png" width=60% height=60%/></center> |
| | <p class="main-page">Since in the beginning the concentration of cAMP is half of all, we can get: | | <p class="main-page">Since in the beginning the concentration of cAMP is half of all, we can get: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/8/89/T--SJTU-BioX-Shanghai--figure1-paper2f.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/8/89/T--SJTU-BioX-Shanghai--figure1-paper2f.png" width=60% height=60%/></center> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | + | |
| | <h3><span class="mw-headline">Reporter protein lightening</span></h3> | | <h3><span class="mw-headline">Reporter protein lightening</span></h3> |
| − | <p class="main-page">As the mechanism of our reporter protein [RRRRR] indicates, we assume that the luminance magnitude is proportional to the concentration of cAMP in the cell, therefore we can get our final formula: | + | <p class="main-page">As the mechanism of our reporter protein [2] indicates, we assume that the luminance magnitude is proportional to the concentration of cAMP in the cell, therefore we can get our final formula: |
| | </p> | | </p> |
| − | <center><img src="/wiki/images/0/0a/T--SJTU-BioX-Shanghai--figure1-paper2g.png" width=100% height=100%/></center> | + | <center><img src="/wiki/images/0/0a/T--SJTU-BioX-Shanghai--figure1-paper2g.png" width=60% height=60%/></center> |
| − | <br>
| + | |
| − | <br>
| + | |
| | <br> | | <br> |
| | <br> | | <br> |
| | <h2><span class="mw-headline">Simulation and results</span></h2> | | <h2><span class="mw-headline">Simulation and results</span></h2> |
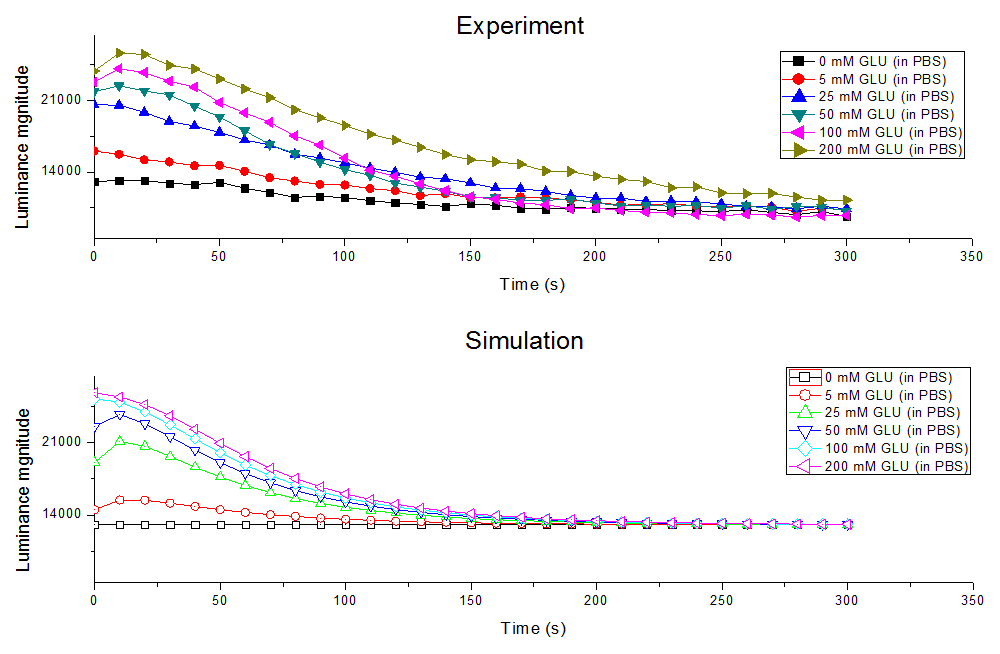
| − | <p class="main-page">We also use the Levenberg-Marguard nonlinear optimization algorithm in this model as numerical simulation | + | <p class="main-page">We also use the Levenberg-Marguard nonlinear optimization algorithm in this model as numerical simulation and get results below. We can see that the figure shows that our simulation is almost the real situation and the <i>f</i> is really small, which means that our model is pretty reliable. The table showed below is the simplified parameters got in our model. The form of which the parameters correspond to the formula showed above is also shown in the table. |
| | </p> | | </p> |
| | + | <center><img src="/wiki/images/a/a9/T--SJTU-BioX-Shanghai--figure1-paper2.png" width=60% height=60%/></center> |
| | + | <center><img src="/wiki/images/5/5a/T--SJTU-BioX-Shanghai--figure1-parameter.PNG" width=30% height=30%/></center> |
| | + | |
| | | | |
| | | | |
| Line 326: |
Line 116: |
| | | | |
| | | | |
| − | <br>
| |
| − | <br>
| |
| | <br> | | <br> |
| | <br> | | <br> |
| | <h2><span class="mw-headline">Reference</span></h2> | | <h2><span class="mw-headline">Reference</span></h2> |
| − | <ul><li>[1] Kholodenko, B. N., Demin, O. V., Moehren, G. & Hoek, J. B. Quantification of short term signaling by | + | <p><i>[1] Kholodenko, B. N., Demin, O. V., Moehren, G. & Hoek, J. B. Quantification of short term signaling by |
| | the epidermal growth factor receptor. J. Biol. Chem.274, 30169–30181 (1999). | | the epidermal growth factor receptor. J. Biol. Chem.274, 30169–30181 (1999). |
| − | </li> | + | </p></i> |
| − | | + | <p><i>[2]Saito K, Chang Y F, Horikawa K, et al. Luminescent proteins for high-speed single-cell and whole-body imaging[J]. Nature communications, 2012, 3: 1262. |
| | + | </i></p> |
| | | | |
| | | | |
| Line 345: |
Line 134: |
| | </p><p><br /> | | </p><p><br /> |
| | </p><p><br /> | | </p><p><br /> |
| − |
| |
| − | <style>
| |
| − | .footer .footer-content {
| |
| − | float: left;
| |
| − | margin-left: 2%;
| |
| − | font-size:80%;
| |
| − | }
| |
| − | .footer p{
| |
| − | color:#f4b045;
| |
| − | }
| |
| − | .footer {
| |
| − | padding-top:8px;
| |
| − | border-top:2px;
| |
| − | border-style:solid none none none;
| |
| − | border-top-color:#f4b045;
| |
| − | margin-bottom:50px;
| |
| − | margin-top:10px;
| |
| − | postion: fixed;
| |
| − | bottom:0;
| |
| − | clear:both;
| |
| − | }
| |
| − |
| |
| − |
| |
| − | </style>
| |
| − | <div class="footer">
| |
| − | <div class="footer-content" style="color:#f4b045;">
| |
| − | <p>Bio-X Institutes</p>
| |
| − | <p>Shanghai Jiao Tong University</p>
| |
| − | <p>800, Dongchuan Road 200240, Shanghai, China</p>
| |
| − | <p>xxxxxxxxx@sjtu.edu.cn</p>
| |
| − | </div>
| |
| − | <div class="footer-logo">
| |
| − | <a href="http://www.sjtu.edu.cn/" title="nima"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/0/03/SJTUB_SJTU_logo.png" height="50" width="150"></a>
| |
| − | <a href="http://www.bio-x.cn/"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/9/95/SJTUB_biox_logo.png" height="50" width="150"></a>
| |
| − | <a href="http://www.sjtu.edu.cn/"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/f/f3/SJTUB_igemfoundation.png" height="50" width="150"></a>
| |
| − | </br>
| |
| − | <a href="http://www.merck.com/index.html"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/2/24/SJTUB_Merk_logo.png" height="50" width="150"></a>
| |
| − | <a href="http://www.genewiz.com/"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/e/e4/SJTUB_genwiz_logo.png" height="50" width="150"></a>
| |
| − | <a href="http://life.sjtu.edu.cn/"><img class="logo_img" border="0" src="https://static.igem.org/mediawiki/2015/0/08/SJTUB_life_logo.png" height="50" width="150"></a>
| |
| − |
| |
| − | </div>
| |
| − | </div>
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − | <style>
| |
| − |
| |
| − | #content {
| |
| − | width: 100%;
| |
| − | }
| |
| − |
| |
| − | article#main {
| |
| − | box-sizing: border-box;
| |
| − | width: 80%;
| |
| − | margin: auto;
| |
| − | padding: 0;
| |
| − |
| |
| − | }
| |
| − | html, body {
| |
| − | background-color: rgba(240, 255, 241, 0.07);
| |
| − | }
| |
| − | p, article#main li{
| |
| − | color: rgb(55, 55, 55);
| |
| − | font: 18px/36px STXihei, sans-serif !important;
| |
| − | vertical-align: middle;
| |
| − | }
| |
| − | #content {
| |
| − | width: 100%;
| |
| − | height: 100%;
| |
| − | box-sizing: border-box;
| |
| − | background-color: transparent;
| |
| − | border: 0px;
| |
| − | }
| |
| − |
| |
| − | .firstHeading {
| |
| − | display: none;
| |
| − | }
| |
| − | h2, h3 {
| |
| − | clear: both;
| |
| − | }
| |
| − |
| |
| − | h2 {
| |
| − | color: #f4b045;
| |
| − | border: 0px;
| |
| − | font-size: 180%;
| |
| − | font-weight: 600;
| |
| − | }
| |
| − |
| |
| − | h3 {
| |
| − | color:#f4b045;
| |
| − | }
| |
| − |
| |
| − | h1 {
| |
| − | color:#f4b045;
| |
| − | }
| |
| − | #toc {
| |
| − | font-size: 0.8em;
| |
| − | }
| |
| − |
| |
| − | figure {
| |
| − | font-size: 0.7em;
| |
| − | }
| |
| − |
| |
| − | .subp {
| |
| − | position: relative;
| |
| − | margin-right: 1.2em;
| |
| − | margin-left: 0.2em;
| |
| − | white-space: nowrap;
| |
| − | }
| |
| − |
| |
| − | .subp > sub {
| |
| − | position: absolute;
| |
| − | top: 50%;
| |
| − | left: 100%;
| |
| − | font-size: 60%;
| |
| − | }
| |
| − |
| |
| − | .subp > sup {
| |
| − | position: absolute;
| |
| − | top: 0%;
| |
| − | left: 100%;
| |
| − | font-size: 60%;
| |
| − | }
| |
| − | @media screen and (min-width:1500px){
| |
| − | #globalWrapper{
| |
| − | font-size:120%;
| |
| − | }
| |
| − | }
| |
| − |
| |
| − |
| |
| − | @media screen and(max-width:1600px) {
| |
| − | #toc{display:none;}
| |
| − | a:link{color:#f4b045;}
| |
| − | #nav nav a {
| |
| − | line-height: 3em;
| |
| − | font-weight: 600;
| |
| − | }
| |
| − | article#main {
| |
| − | margin-right: 0px;
| |
| − | margin-left: 20%;
| |
| − | width: auto;
| |
| − | }
| |
| − | a, a:link, a:visited {color: #f4b045;}
| |
| − | }
| |
| − |
| |
| − | a, a:link, a:visited {color: #f4b045;}
| |
| − |
| |
| − | </style>
| |
| − |
| |
| − | <script>
| |
| − | $(document).ready(function(){$(".mw-body a.external").css("color", "#f4b045");})
| |
| − |
| |
| − | </p><p><style>
| |
| − | article#main {
| |
| − | width: 80%;
| |
| − | margin-left:15%;
| |
| − | padding: 0;
| |
| − | }
| |
| − |
| |
| − | p,article#main li, {
| |
| − | margin-left:10%;
| |
| − | width:80%;
| |
| − | color: rgb(55, 55, 55);
| |
| − | font-family: raleway-regular, sans-serif;
| |
| − | line-height: 1.7em;
| |
| − | }
| |
| − |
| |
| − | h1 {
| |
| − | margin-left: 7%;
| |
| − | font-weight: 800;
| |
| − | border: 0px;
| |
| − | padding-bottom:none;
| |
| − | padding-top: 0.5em;
| |
| − | color: #f4b045;
| |
| − | cursor: auto;
| |
| − | font-family:raleway-regular, sans-serif;
| |
| − | font-size: 3.5em;
| |
| − | line-height:1;
| |
| − | font-style: normal;
| |
| − | font-variant: normal;
| |
| − | }
| |
| − | #globalWrapper {
| |
| − | padding:0;
| |
| − | margin:0;
| |
| − | font-size:100%;
| |
| − | }
| |
| − | body{
| |
| − | font: initial;
| |
| − | }
| |
| − | </style>
| |
| − |
| |
| − | </p>
| |
| | | | |
| | | | |
| Line 548: |
Line 143: |
| | </body> | | </body> |
| | </html> | | </html> |
| | + | {{SJTU-BioX-Shanghai/Footer}} |
| | + | {{SJTU-BioX-Shanghai/Style}} |