(→Mass spectroscopy) |
(→Measurement) |
||
| (30 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
{{LMU-TUM_Munich|navClass=project}} | {{LMU-TUM_Munich|navClass=project}} | ||
__NOTOC__ | __NOTOC__ | ||
| + | <div class="white-box"> | ||
| + | [[File:Muc16_BioBrick_summary_001.png |frameless|center|820px]] | ||
| + | </div> | ||
| − | + | =The sub-projects of biotINK and their achievements= | |
| − | = | + | |
<div class="white-box"> | <div class="white-box"> | ||
| − | |||
| − | |||
| − | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Design BioBrick Part Design] == | |
| + | [[File:Muc16_Sticker_Design_001.png |right|200px]] | ||
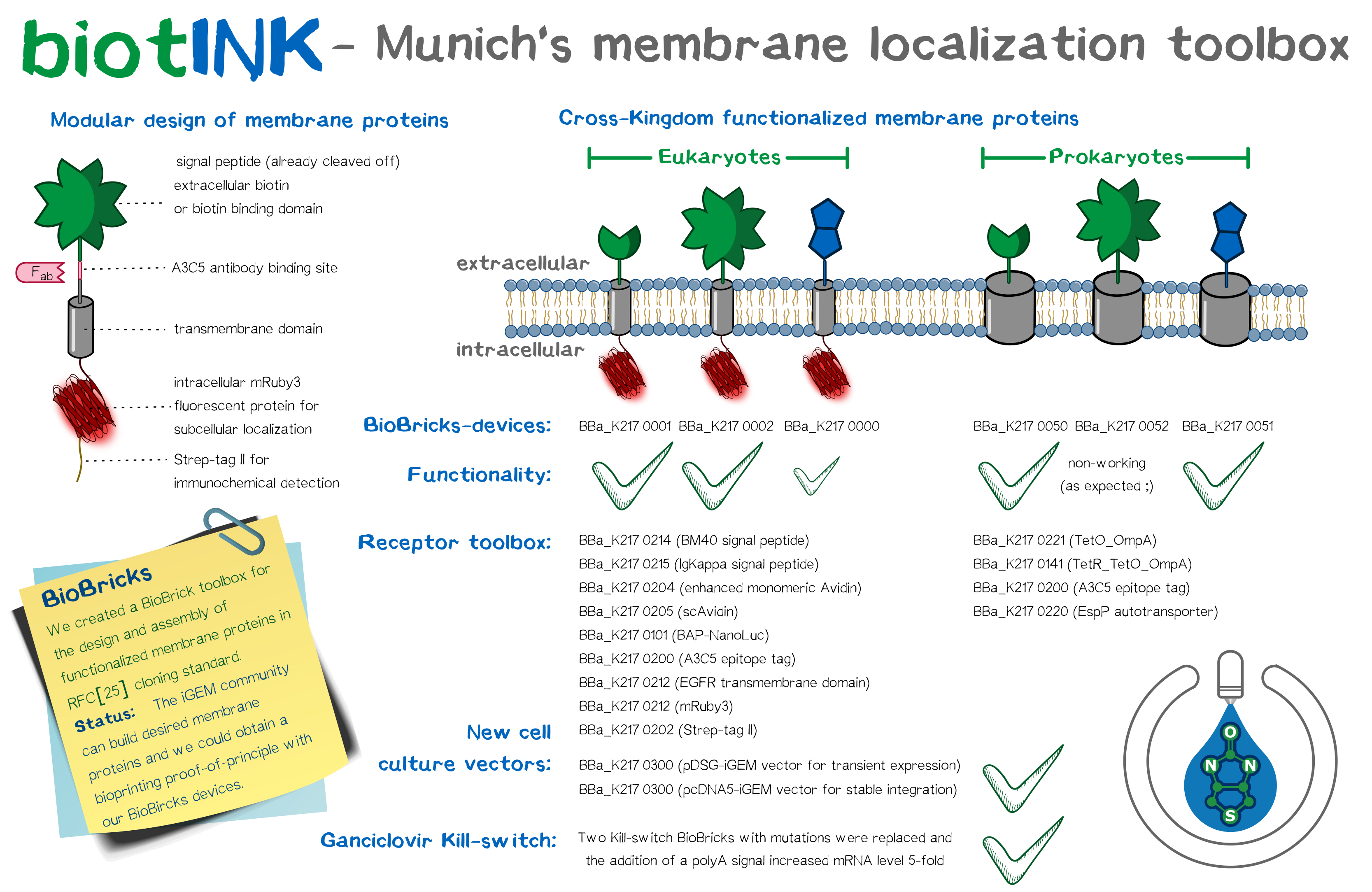
| + | As does every sophisticated SynBio project, we designed a plethora of BioBrick constructs and devices in the course of our project. Most prominent among these are the modular transmembrane proteins we conceptualized, since they are integral to the functionality of our bioprinting system. Basing our workflow on the standard RFC[10] and RFC[25] assembly standards, we assembled the same intracellular and transmembrane domains, namely mRuby3 and the EGFR transmembrane domain, with different extracellular domains, among them single-chain avidin, monomeric avidin, and NanoLuc fused with an N-terminal Avi-tag for ''in vivo'' biotinylation by the co-expressed biotin ligase BirA. We coupled these constructs with a trafficking signal for membrane integration. Including further elements to stabilize these receptors offers advantages over previously assembled BioBricks, and planning our constructs with multiple options for detection provided us ample opportunity to extensively characterize the outcome. Apart from these eukaryotic designs, we drafted an autotransporter construct for bacterial surface display. | ||
| + | <br><br><br> | ||
| − | == | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Proteins Production, purification & conjugation of recombinant proteins] == |
| − | + | [[File:Muc16_Sticker_proteins_001.png |right|200px]] | |
| + | As the second constituent of our two-component polymerization system next to the membrane proteins, we required large amounts of protein, either biotin-binding or bearing chemically introduced biotin depending on the receptor in question. For biotin-binding proteins, we produced and purified the avidin variants wt streptavidin; traptavidin,a variant with higher affinity toward biotin and enhanced thermal stability; streptactin, a variant that binds an oligopeptide sequence termed strep-tag; and enhanced monomeric avidin, which does not assemble into tetramers and is thus monovalent. wt Streptavidin was produced both in the dimensions of shaker flasks and a fermenter, and purified according to an established protocol. As a scaffold for chemical biotinylation, we produced and purified PAS-lysine and GFP. We extensively characterized all proteins as well as the biotinylated forms of the latter group plus BSA by ESI-TOF, SDS-PAGE, and IEF; moreover, we determined the affinity of streptavidin variants ''via'' fluorescence titration and their thermal stability using CD spectra. We produced all target proteins in good purity and successfully biotinylated PAS-lysine, GFP, and BSA. | ||
| + | <br><br><br><br><br> | ||
| − | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Receptors A fully functional cross-kingdom membrane localization toolbox] == | |
| − | + | [[File:Muc16_Sticker_Receptors_003.png |right|200px]] | |
| − | File: | + | Based on our modular membrane protein design, we evaluated the functionality of different signal peptide in shuttling the protein into the membrane. Apart from taking the BioBrick EGFR signal peptide, we designed two further signal peptides with increased translational efficiency, incorporating items such as the Kozak sequence. These three signal peptides were analyzed ''in silico'' as well as experimentally using a luciferase secretion assay. The BM40 signal, one of our self-designed constructs, was most efficient and thus incorporated into our other devices. Additionally, we incorporated three reporter modules (mRuby3, A3C5 epitope tag and strep-tag II) into the receptors. The finished constructs were used to generate semi-stablly transfected and stable cell lines. Protein localization was confirmed ''via'' fluorescence and high-resolution fluorescence microscopy, including characterization of a trafficking signal for increased membrane localization, and the recombinant protein was quantified by FACS and Western blot. mRNA expression levels were detected by RT-q-PCR. For visualization, aggregation of functionalized cells with Strep beads was subjected to SEM. The bacterial autotransporter was characterized and found to successfully shuttle small protein domains to the extracellular surface. |
| − | + | <br><br><br><br><br> | |
| − | < | + | |
| − | == | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Linkerchemistry Chemical synthesis of amino-reactive biotinylation compounds] == |
| − | + | [[File:Muc16_Sticker_Chemistry_001.png |right|200px]] | |
| − | + | Apart from the standard biotin-NHS, several other derivatives spacing biotin and NHS apart by an amino acid residue were synthesized. These derivatives were used in the chemical biotinylation of BSA in order to assess their relative stability toward cleavage by the plasma protein biotinidase. The assay principle is based on a colorimetric reaction and was developed by ourselves. Compared to biotinylation ''via'' regular biotin-NHS, the derivative incorporating a spacer aminocaproic acid is cleaved somewhat faster, allowing to dissolve a cell mesh more quickly, and the spacer valine resulted in a drastically slowed hydrolysis rate. | |
| − | + | <br><br><br><br><br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | == | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Proof Proof of principle for the avidin-biotin "polymerization"] == |
| − | + | [[File:Muc16_Sticker_Proof_001.png |right|200px]] | |
| − | + | We chose the avidin-biotin system because of their high affinity and rapid association. Our successful polymerization tests showed that cell density, concentration of biotinylated BSA in the bioink, and streptavidin concentration in the reservoir are the three major factors that impact performance. Thus, more BSA entailed higher structural integrity. We successfully printed defined objects with our technology and developed an optimal ratio of constituents. Aggregation was visually confirmed in the micromanipulator. | |
| + | <br><br><br><br><br> | ||
| − | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Demonstrate First bioprinting results using the biotINK bioprinter] == | |
| − | + | [[File:Muc16_Sticker_Demonstrate_001.png |right|200px]] | |
| − | + | We successfully produced first prints of defined spatial structure in our bioprinter and show that our technology compares favorably with existing Inkjet and microextrusion technologies in bioprinting, especially in terms of cell viability. | |
| − | + | <br><br><br><br><br> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Hypoxia A vascularization inducer based on a hypoxia-sensitive promoter] == | |
| − | + | [[File:Muc16_Sticker_Hypoxia_001.png |right|200px|]] | |
| − | + | We designed a hypoxia-driven promoter, which we tested for its functionality under low oxygen as well as normal oxygen conditions. Although the promoter does not increase gene expressions when cells are exposed to hypoxia, it remains remarkably steady - a feature that most promoters do not have. | |
| − | + | ||
| − | + | <br><br><br><br><br> | |
| − | + | ||
| − | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Safety Safety evaluation of our project and the kill-switch] == | |
| − | == | + | [[File:Muc16_Sticker_Safety_001.png |right|200px|]] |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | <br> | |
| − | + | ||
| − | < | + | |
| − | + | Sequencing results showed a point mutation in both the part by the iGEM team of Freiburg 2010 and the Slovenian part of 2012. The mutation led to a PstI restriction site, that complicated cloning according to iGEM standards. After a site-directed mutagenesis (Quickchange) the restriction site was removed and the sequence hence repaired. <br> | |
| − | < | + | Quantitative analysis by mRNA measuring of Poly-A-sequence addition showed 4.6-fold higher mRNA stability in comparison to the non-Poly-A-construct. <br> |
| − | < | + | Kill-Switch induction turned out to be difficult when using the prodrug famciclovir. Although HEK293T cells are known to produce aldehyde oxidase <ref> Moriwaki, Y., Yamamoto, T., Takahashi, S., Tsutsumi, Z., & Hada, T. (2001). Widespread cellular distribution of aldehyde oxidase in human tissues found by immunohistochemistry staining. Histology and histopathology, 16(3), 745-753. </ref> and xanthine oxidase <ref> Czupryna, J., & Tsourkas, A. (2012). Xanthine oxidase‐generated hydrogen peroxide is a consequence, not a mediator of cell death. FEBS Journal, 279(5), 844-855. </ref> the conversion of famciclovir into its active form penciclovir seems not to proceed effectively enough in HEK293T cells. |
| − | </ | + | |
| − | == | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/InterLab '''InterLab study''': Determining reliability of measurements] == |
| + | [[File:Muc16_Sticker_Interlab_001.png |right|200px|]] | ||
| − | + | We participated in this year's interlab study and measured five GFP expression plasmids in 8 different ''E. coli'' strains. In addition of measuring according to the standard protocol, we were able to improve upon the protocol in order to make measurements more comparable and allow the analysis of more strains at once. | |
| − | + | ||
| − | = | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Entrepreneurship Entrepreneurship] == |
| + | [[File:Muc16_Sticker_Entrepreneurship_001.png |right|200px|]] | ||
| − | + | For the entrepeneurial part of our project, we firstly created a business-plan for a possible commercial cause. Secondly, we took a look at enabling factors for startups resulting from iGEM via a survey, and analyzed what makes iGEM projects become a startup. | |
| + | <br><br><br><br><br> | ||
| − | = | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Hardware Hardware] == |
| + | [[File:Muc16_Sticker_Hardware_001.png |right|200px|]] | ||
| + | We were able to refunctionalize a conventional 3D-printer by replacing the plastic printing head with a syringe pump. For that purpose, we were able to print most of the new parts of the bioprinter via the 3D-printer itself. The low costs of rebuilding the printer allows the design to be adapted easily by other labs as well, thus allowing other groups to easily adapt our technique.<br><br> | ||
| − | = | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Software Software] == |
| + | [[File:Muc16_Sticker_Software_001.png |right|200px]] | ||
| + | Since the Ultimaker 2+ standard software is not versatile enough to allow for all requirements of bioprinting, we modified the software code in order to make the printing system more adaptable and perfectly suited to the needs of printing live cells. A number of parameters can now be specifically addressed and are tailored to bioprinting. | ||
| + | <br><br><br><br><br> | ||
| − | = | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Measurement Measurement] == |
| + | [[File:Muc16_Sticker_Measurement_001.png |right|200px]] | ||
| + | <br> | ||
| + | The majority of our measurements focussed on the characterization of procaryotic and eucaryotic membrane proteins and includes various techniques. We could confirm the functionality of four biobrick devices and identify mRNA stability as a crucial factor for high surface presentation of membrane proteins. | ||
| − | = | + | == [https://2016.igem.org/Team:LMU-TUM_Munich/Model Model] == |
| − | + | [[File:Muc16_Sticker_Model_001.png |right|200px]] | |
| − | = | + | In our modelling project we have developed a mathematical model for the polymerization of multivalent proteins and cells. |
| − | < | + | </div> |
{{LMU-TUM_Munich_html_end}} | {{LMU-TUM_Munich_html_end}} | ||
Latest revision as of 03:52, 20 October 2016
The sub-projects of biotINK and their achievements
BioBrick Part Design
As does every sophisticated SynBio project, we designed a plethora of BioBrick constructs and devices in the course of our project. Most prominent among these are the modular transmembrane proteins we conceptualized, since they are integral to the functionality of our bioprinting system. Basing our workflow on the standard RFC[10] and RFC[25] assembly standards, we assembled the same intracellular and transmembrane domains, namely mRuby3 and the EGFR transmembrane domain, with different extracellular domains, among them single-chain avidin, monomeric avidin, and NanoLuc fused with an N-terminal Avi-tag for in vivo biotinylation by the co-expressed biotin ligase BirA. We coupled these constructs with a trafficking signal for membrane integration. Including further elements to stabilize these receptors offers advantages over previously assembled BioBricks, and planning our constructs with multiple options for detection provided us ample opportunity to extensively characterize the outcome. Apart from these eukaryotic designs, we drafted an autotransporter construct for bacterial surface display.
Production, purification & conjugation of recombinant proteins
As the second constituent of our two-component polymerization system next to the membrane proteins, we required large amounts of protein, either biotin-binding or bearing chemically introduced biotin depending on the receptor in question. For biotin-binding proteins, we produced and purified the avidin variants wt streptavidin; traptavidin,a variant with higher affinity toward biotin and enhanced thermal stability; streptactin, a variant that binds an oligopeptide sequence termed strep-tag; and enhanced monomeric avidin, which does not assemble into tetramers and is thus monovalent. wt Streptavidin was produced both in the dimensions of shaker flasks and a fermenter, and purified according to an established protocol. As a scaffold for chemical biotinylation, we produced and purified PAS-lysine and GFP. We extensively characterized all proteins as well as the biotinylated forms of the latter group plus BSA by ESI-TOF, SDS-PAGE, and IEF; moreover, we determined the affinity of streptavidin variants via fluorescence titration and their thermal stability using CD spectra. We produced all target proteins in good purity and successfully biotinylated PAS-lysine, GFP, and BSA.
A fully functional cross-kingdom membrane localization toolbox
Based on our modular membrane protein design, we evaluated the functionality of different signal peptide in shuttling the protein into the membrane. Apart from taking the BioBrick EGFR signal peptide, we designed two further signal peptides with increased translational efficiency, incorporating items such as the Kozak sequence. These three signal peptides were analyzed in silico as well as experimentally using a luciferase secretion assay. The BM40 signal, one of our self-designed constructs, was most efficient and thus incorporated into our other devices. Additionally, we incorporated three reporter modules (mRuby3, A3C5 epitope tag and strep-tag II) into the receptors. The finished constructs were used to generate semi-stablly transfected and stable cell lines. Protein localization was confirmed via fluorescence and high-resolution fluorescence microscopy, including characterization of a trafficking signal for increased membrane localization, and the recombinant protein was quantified by FACS and Western blot. mRNA expression levels were detected by RT-q-PCR. For visualization, aggregation of functionalized cells with Strep beads was subjected to SEM. The bacterial autotransporter was characterized and found to successfully shuttle small protein domains to the extracellular surface.
Chemical synthesis of amino-reactive biotinylation compounds
Apart from the standard biotin-NHS, several other derivatives spacing biotin and NHS apart by an amino acid residue were synthesized. These derivatives were used in the chemical biotinylation of BSA in order to assess their relative stability toward cleavage by the plasma protein biotinidase. The assay principle is based on a colorimetric reaction and was developed by ourselves. Compared to biotinylation via regular biotin-NHS, the derivative incorporating a spacer aminocaproic acid is cleaved somewhat faster, allowing to dissolve a cell mesh more quickly, and the spacer valine resulted in a drastically slowed hydrolysis rate.
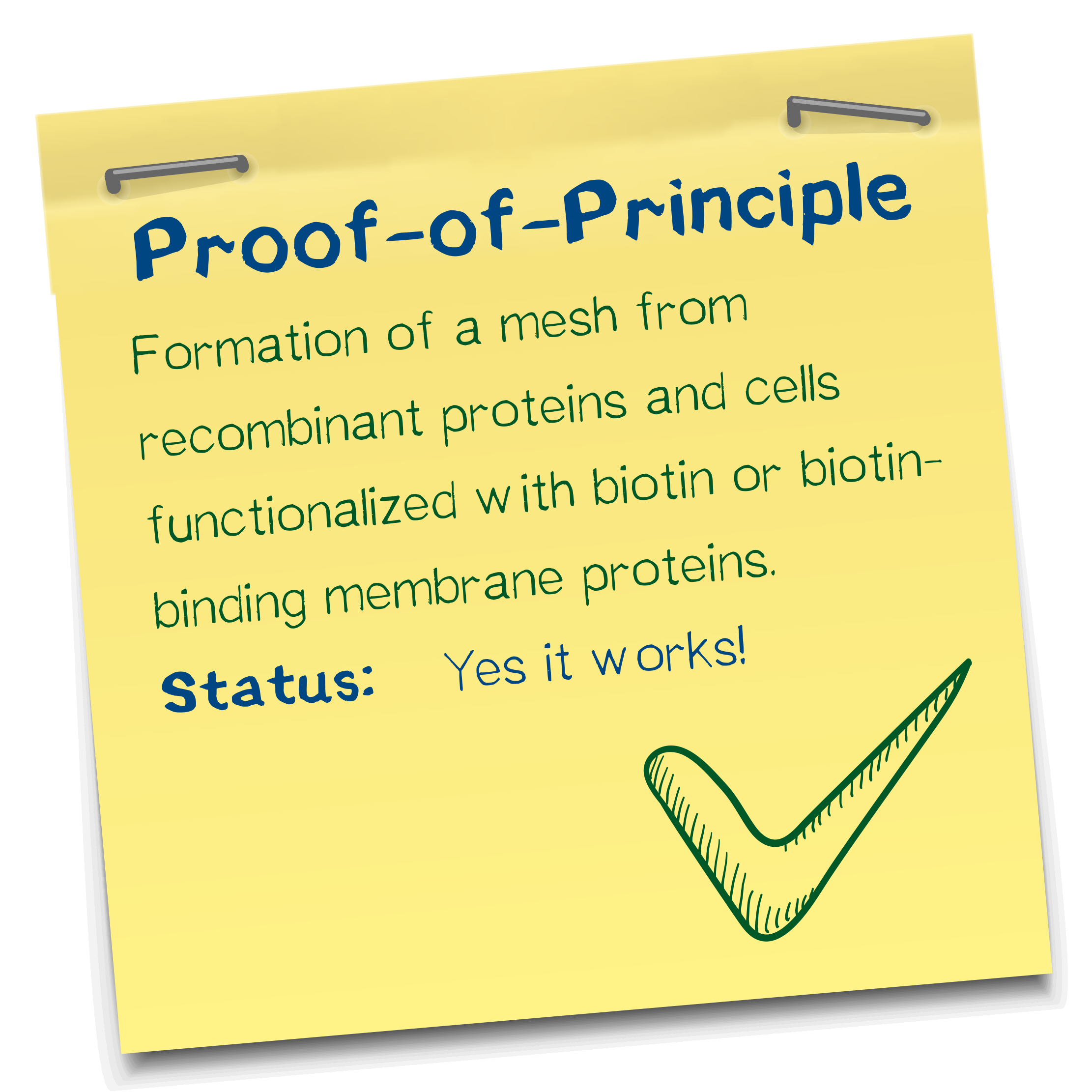
Proof of principle for the avidin-biotin "polymerization"
We chose the avidin-biotin system because of their high affinity and rapid association. Our successful polymerization tests showed that cell density, concentration of biotinylated BSA in the bioink, and streptavidin concentration in the reservoir are the three major factors that impact performance. Thus, more BSA entailed higher structural integrity. We successfully printed defined objects with our technology and developed an optimal ratio of constituents. Aggregation was visually confirmed in the micromanipulator.
First bioprinting results using the biotINK bioprinter
We successfully produced first prints of defined spatial structure in our bioprinter and show that our technology compares favorably with existing Inkjet and microextrusion technologies in bioprinting, especially in terms of cell viability.
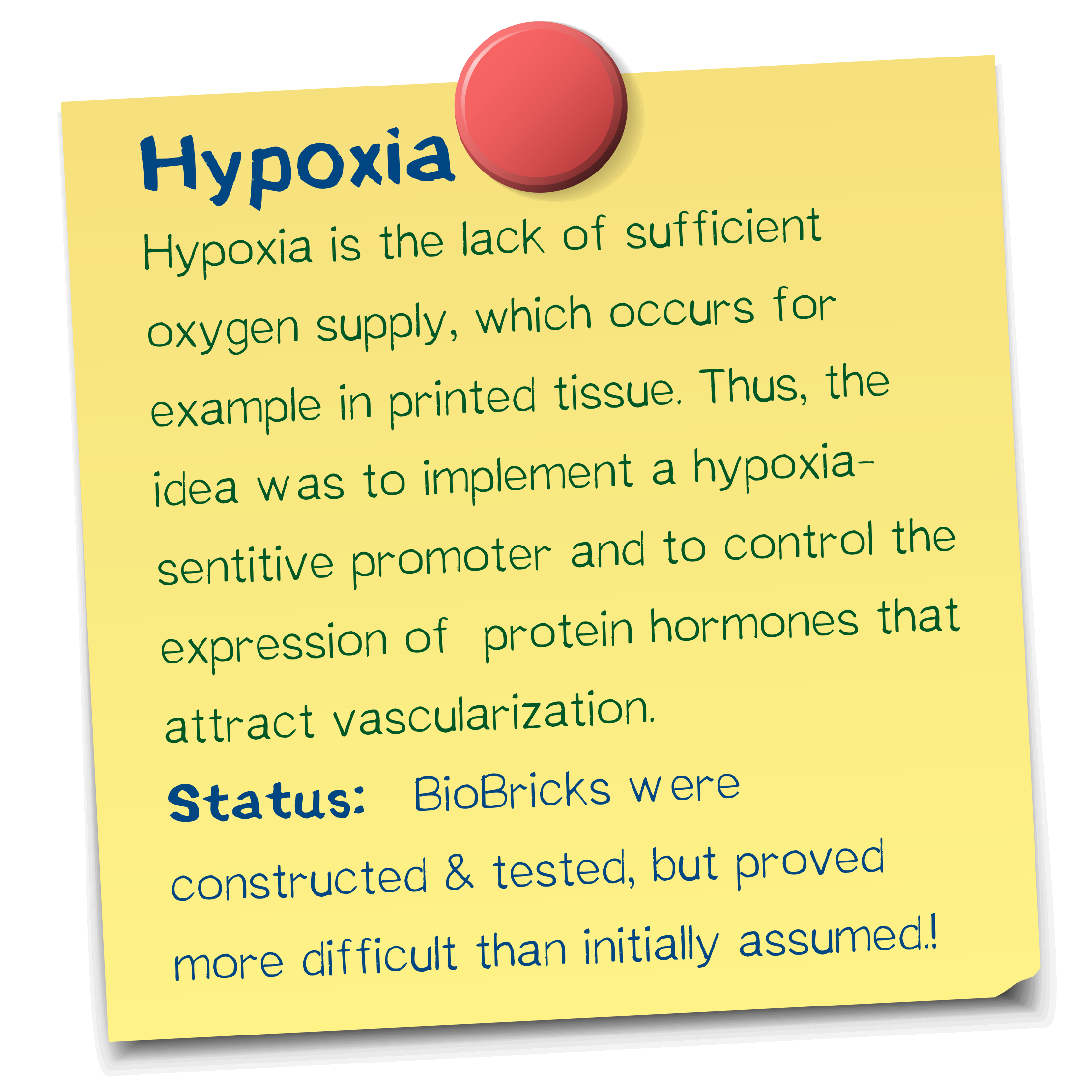
A vascularization inducer based on a hypoxia-sensitive promoter
We designed a hypoxia-driven promoter, which we tested for its functionality under low oxygen as well as normal oxygen conditions. Although the promoter does not increase gene expressions when cells are exposed to hypoxia, it remains remarkably steady - a feature that most promoters do not have.
Safety evaluation of our project and the kill-switch
Sequencing results showed a point mutation in both the part by the iGEM team of Freiburg 2010 and the Slovenian part of 2012. The mutation led to a PstI restriction site, that complicated cloning according to iGEM standards. After a site-directed mutagenesis (Quickchange) the restriction site was removed and the sequence hence repaired.
Quantitative analysis by mRNA measuring of Poly-A-sequence addition showed 4.6-fold higher mRNA stability in comparison to the non-Poly-A-construct.
Kill-Switch induction turned out to be difficult when using the prodrug famciclovir. Although HEK293T cells are known to produce aldehyde oxidase [1] and xanthine oxidase [2] the conversion of famciclovir into its active form penciclovir seems not to proceed effectively enough in HEK293T cells.
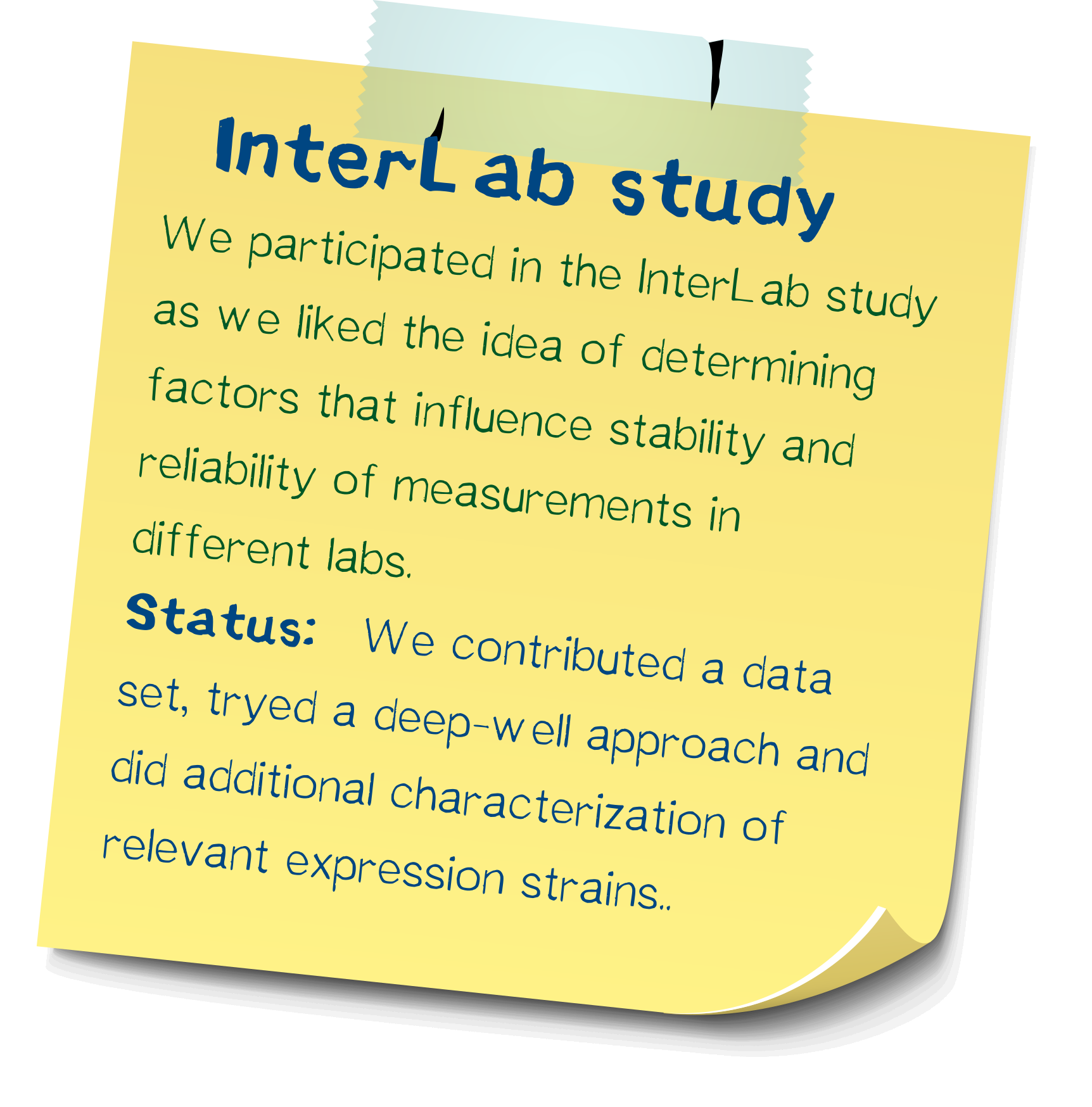
InterLab study: Determining reliability of measurements
We participated in this year's interlab study and measured five GFP expression plasmids in 8 different E. coli strains. In addition of measuring according to the standard protocol, we were able to improve upon the protocol in order to make measurements more comparable and allow the analysis of more strains at once.
Entrepreneurship
For the entrepeneurial part of our project, we firstly created a business-plan for a possible commercial cause. Secondly, we took a look at enabling factors for startups resulting from iGEM via a survey, and analyzed what makes iGEM projects become a startup.
Hardware
We were able to refunctionalize a conventional 3D-printer by replacing the plastic printing head with a syringe pump. For that purpose, we were able to print most of the new parts of the bioprinter via the 3D-printer itself. The low costs of rebuilding the printer allows the design to be adapted easily by other labs as well, thus allowing other groups to easily adapt our technique.
Software
Since the Ultimaker 2+ standard software is not versatile enough to allow for all requirements of bioprinting, we modified the software code in order to make the printing system more adaptable and perfectly suited to the needs of printing live cells. A number of parameters can now be specifically addressed and are tailored to bioprinting.
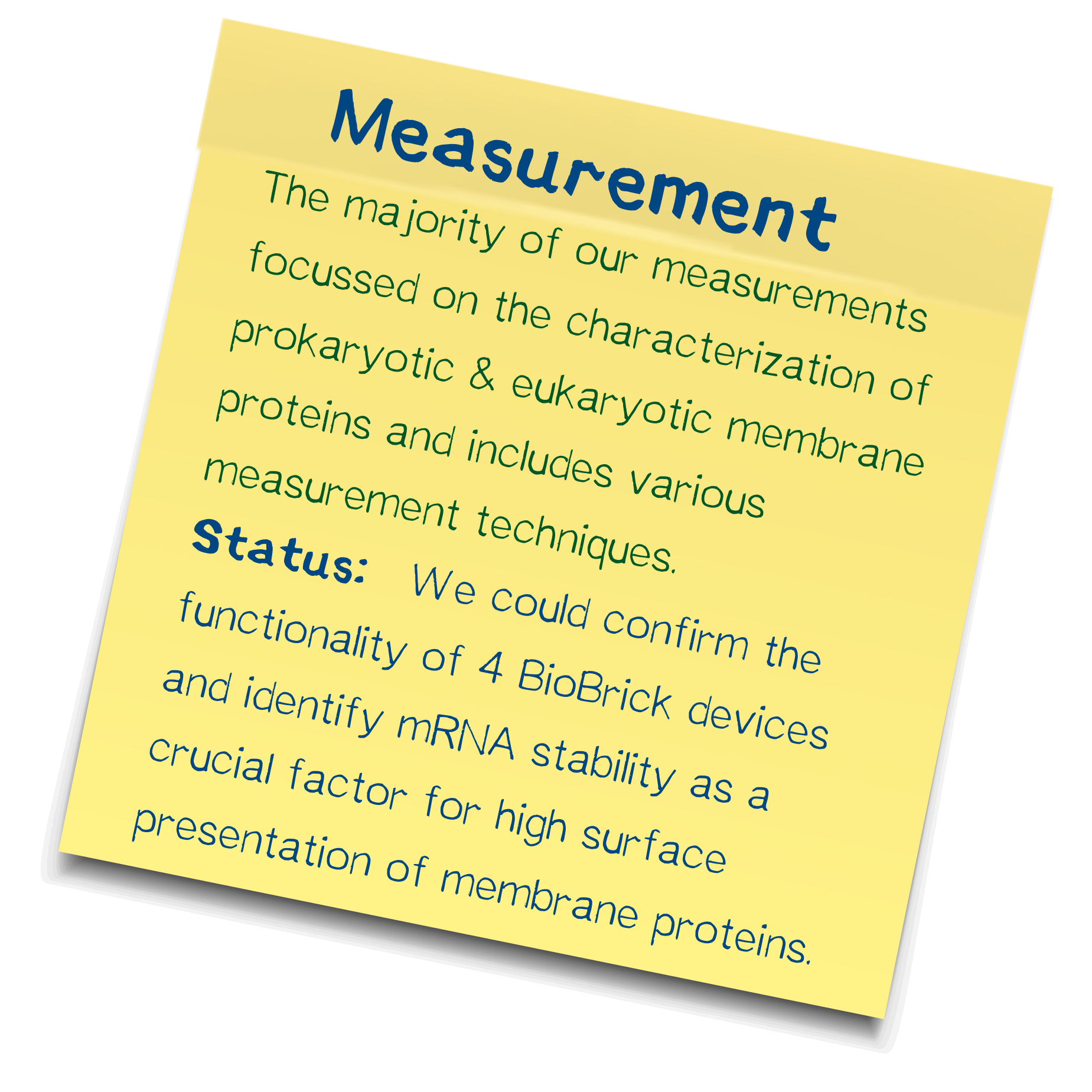
Measurement
The majority of our measurements focussed on the characterization of procaryotic and eucaryotic membrane proteins and includes various techniques. We could confirm the functionality of four biobrick devices and identify mRNA stability as a crucial factor for high surface presentation of membrane proteins.
Model
In our modelling project we have developed a mathematical model for the polymerization of multivalent proteins and cells.
- ↑ Moriwaki, Y., Yamamoto, T., Takahashi, S., Tsutsumi, Z., & Hada, T. (2001). Widespread cellular distribution of aldehyde oxidase in human tissues found by immunohistochemistry staining. Histology and histopathology, 16(3), 745-753.
- ↑ Czupryna, J., & Tsourkas, A. (2012). Xanthine oxidase‐generated hydrogen peroxide is a consequence, not a mediator of cell death. FEBS Journal, 279(5), 844-855.